
A Python SDK for Authoring and using Computer-interpretable
Guidelines
Marcus Barann, Stefan Heldmann
a
, Jan Klein
b
and Stefan Kraß
Fraunhofer Institute for Digital Medicine MEVIS, Bremen/L
¨
ubeck, Germany
Keywords:
Computer-interpretable Guidelines, Decision Support Systems.
Abstract:
In this paper we describe a Python SDK that we developed and which we used to create a decision support
system (DSS) for determining and presenting clinical practice guideline (CPG) recommendations for individ-
ual patients. Computer-interpretable guidelines (CIGs) are formalisms that represent CPG knowledge. Our
Python SDK implements a model and an engine for a CIG formalism that can be easily integrated into any
Python-based application. We describe important aspects of creating a guideline model with our CIG and
present a web application that interacts with our guideline engine through a REST API. The web application
implements generic components to manage and display the current input needs, recommendations, and state-
ments. In comparison to PROforma, we added predicate components, which facilitate the reuse of logical
expressions. Arguments refer to predicates instead of including expressions. This allows reusing the same
expression in multiple arguments. We also allow the use of the predicates in other expressions, like in expres-
sions of other predicates and task preconditions. To facilitate the integration of our CIG in decision support
systems, we added properties to all PROforma components that represent a code from a terminology system.
1 INTRODUCTION
Clinical research aims at improving effectiveness and
efficiency of patient health care. The increasing
research generates a growing amount of evidence.
Therefore, it is difficult for practitioners to keep up
with the most recent knowledge about the best patient
treatment. To overcome this, groups of clinical ex-
perts study and evaluate new evidence and formulate
recommendations and statements for the patient treat-
ment. They publish their results in form of clinical
practice guidelines (CPGs) (?).
These written CPGs target a specific area of health
care, for example, the treatment of a specific disease
that occurs in a defined population. During a patient
encounter, a practitioner must identify the parts of the
guideline that apply to the patient, because guidelines
refer generally to a population. For a single patient
only specific parts of a CPG are of relevance. CPGs
can be quite comprehensive documents, so this can
be a tedious task. This is even more likely for pa-
tients with comorbidities, where multiple CPGs need
to be consulted. Additionally, they might contradict
a
https://orcid.org/0000-0002-9206-2086
b
https://orcid.org/0000-0002-0881-408X
each other, which would have to be recognized and
resolved. As a solution to this, computer-interpretable
guidelines (CIGs) have been invented. They are for-
malisms with concepts to represent the CPG knowl-
edge (?; ?). By integrating CIG models into decision
support systems, and allowing them to access infor-
mation in patient health records (PHRs), the CIGs can
identify CPG knowledge that is relevant for an indi-
vidual patient. This happens in real-time and can be
integrated into the practitioners daily routine. Deci-
sion support systems (DSS) could also include differ-
ent CIGs and automatically manage contradictions to
support the care of patients with multiple or comor-
bidities (?).
Although computer-interpretable guidelines have
been researched for decades now, it has not been es-
tablished that they are widely used in clinical routine.
Instead, CPGs are still the common form for transfer-
ring evidence-based knowledge into the clinical rou-
tine. A reason for this might be that the transforma-
tion of a CPG into a CIG is often a time-consuming
and coordination-intensive process, as expert knowl-
edge is needed from two domains, the clinical and the
technological area.
Another reason could be the low availability
of standardized and user-friendly tools for creating
Barann, M., Heldmann, S., Klein, J. and Kraß, S.
A Python SDK for Authoring and using Computer-interpretable Guidelines.
DOI: 10.5220/0010792800003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 3: BIOINFORMATICS, pages 99-106
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
99

CIGs. While several tools exist that support writ-
ing CPG text documents and publishing them as PDF
documents (Microsoft Word, LibreOffice, Docbook,
LaTeX), editors for CIGs are rare. The ontology ed-
itor Prot
´
eg
´
e (?) has been used as visual editor for
models of different CIG formalisms (?, p. 751) and
AsbruView is used to create models of the Asbru for-
malism (?). The PROforma formalism also has its
own tools, which are Tallis and Alium from Deontics
(?), and Arezzo from Elsevier InferMed (?). Arezzo
and Alium are commercial tools, while Tallis (?) is
available for research only.
Khodambashi et al. performed a review and anal-
ysis in the field of CIGs (?). They evaluated aspects of
the following categories: “knowledge extraction per-
formance, tool functionality, modelling perspective,
user’s effect on modelling, didactic support, usabil-
ity, guideline verification and validation, integration,
maintenance features and comparative analysis on en-
coding process”. They concluded that “mainly techni-
cal and functional aspects of the technology” are ad-
dressed in the field of CIGs and postulated that it has
“to move to a stronger focus on its users”. We also
believe that a strong focus on users is important, but
the complexity of modelling a CIG cannot be avoided.
Analytic and process oriented thinking is necessary,
regardless of the tooling.
Furthermore, a CIG needs access to data from a
PHR. Until today, accessing data from a PHR can im-
pose a high effort, which can be an obstacle in the
development of a CIG integrating DSS.
First, if digitally structured data is available, it is
often not accessible through a standardized interface.
This increases the effort to install a DSS with an in-
tegrated CIG in a clinical environment. However, the
open HL7 FHIR standard (?) is being developed and
increasingly used. It provides a standardized inter-
face to access data from PHRs, reducing the effort to
install decision support systems in different clinical
environments.
Second, data is often stored unstructured and in
natural language, which makes data processing diffi-
cult or even impossible. This means that a DSS using
a CIG can not automatically access data, but requires
a user to input it. This counteracts the time saving
benefit of using a CIG over a CPG, especially with an
increasing amount of data. We think that solutions to
inquire digitally structured data need to be developed,
because an automated processing of unstructured data
in natural languange will not be generally possible.
We believe that more and more structured data
will be available through a standardized interface in
the near future, increasing the applicability of CIGs.
This motivated us to find a way to apply clinical
guidelines to patient data that is stored in a database.
For the interpretation of patient data, medical termi-
nologies like UMLS or SNOMED CT are important.
We developed a software framework for authoring
CIG models, into which we could integrate the sup-
port of such terminologies to facilitate semantically
joining CIG components and patient data. In order to
be able to reuse the technology, we decided to develop
it as a Python SDK rather than a monolithic applica-
tion.
2 METHODS
Several CIG formalisms have been introduced over
the last two decades (?; ?; ?). M. Peleg (?) di-
vides CIG formalisms into three categories: docu-
ment models, decision trees and probabilistic mod-
els, and task-network models. Task-network mod-
els (TNMs) “provide modeling primitives specifically
designed for representing complex, multistep clinical
guidelines and for describing temporal and other re-
lationships between component tasks. Unlike rule-
based systems, TNMs can explicitly model alternative
pathways or sequences of tasks (i.e., control flow),
and they provide tools for visual representation of
plans and the organization of tasks within them” (?,
p. 53).
Especially GLIF (?) and PROforma (?; ?), two
task-network model formalisms, have shown a high
relevance. For our development, we selected PRO-
forma as formalism as it is used in several research
projects (?; ?; ?; ?; ?; ?; ?; ?; ?; ?; ?) as well as
in commercial solutions (?). From this we concluded
that PROforma has the potential of being a universal
CIG formalism and that it should fulfil our needs.
2.1 Implementation of PROforma
Concepts
We implemented the PROforma concepts and guide-
line engine in the Python programming language. A
detailed description of PROforma is given in (?). We
chose Python, because it is an interpreted language
that allows to evaluate expressions during runtime.
This saved us the effort to implement an own expres-
sion evaluator. Python’s popularity in the scientific
community (?) additionally influenced our choice.
The PROforma components have been imple-
mented as Python classes. They include the compo-
nent data and parameterization. In our implementa-
tion the components have property objects that store
values and changes locally. This is a difference to
the original PROforma formalism, which has a global
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
100
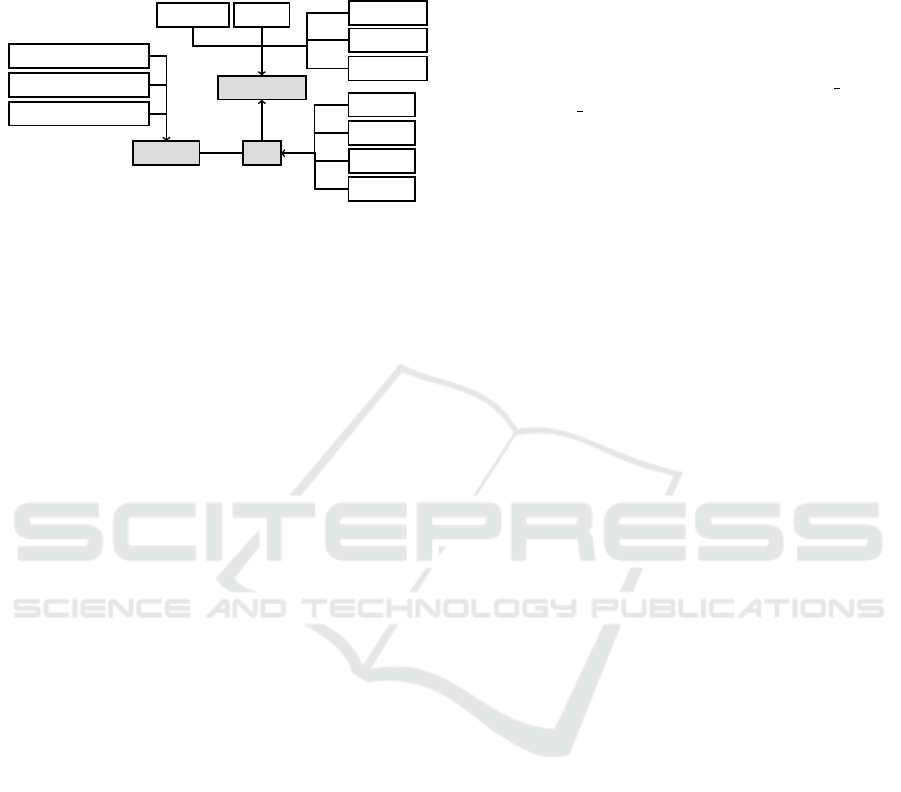
properties table that stores the component data and a
global changes tables that stores value changes. Fig-
ure ?? shows the hierarchy of the most central classes.
Component
Action
Task
Property
Decision
Enquiry
Plan
DataItem
Source
ExpressionProperty
NumericProperty
StringProperty
Predicate
Argument
Candidate
1
*
Figure 1: This is the hierarchy of our Python classes
that implement the PROforma components and properties.
The base classes Component, Task, and Property are high-
lighted. One Task class contains multiple Property classes.
For each component we implemented a compo-
nent controller class, which implements the engine
logic and separates it from the component data. For
example, the class TaskController can start or discard
a task.
We do not parse and evaluate the original PRO-
forma syntax. Our guideline models are persisted as
RDF (?) documents using the turtle syntax (?). This
saved us the effort to implement a parser for the PRO-
forma syntax. The ontology of our guideline formal-
ism is also available as an RDF document, which is
automatically generated from the Python classes.
Logical expressions are written in Python syntax.
They are allowed to use only a defined set of functions
instead of the entire Python API to avoid adding arbi-
trary side effects and to reduce the risk of injecting
malicious code through guideline models into a DSS.
In comparison to PROforma, we extended our
CIG as follows:
• We added predicate components, which facilitate
the reuse of logical expressions. Decision candi-
date arguments refer to a predicate instead of in-
cluding an expression. This allows for reusing the
same expression in multiple arguments by refer-
ring to the same predicate. We also support the
use of predicates in other expressions, like the
expressions of other predicates and task precon-
ditions. Reusing expressions corresponds to the
software development practice to prevent code du-
plication through modularization, which improves
code maintainability. This makes expressions less
complex and further corrections of a predicate
will then automatically affect all expressions that
use it. This saves the author time and prevents er-
rors, because no manual search for and correction
of the otherwise recurring pattern in expressions
is necessary.
• To facilitate the integration of our CIG in decision
support systems, we added properties to all com-
ponents that can represent a code from a terminol-
ogy system. For example, the candidates of a de-
cision can be labelled by codes from the “Unified
Medical Language System” (UMLS) ontology.
The properties, inspired by the Coding element
of the FHIR standard (?), are: code, code system,
and code display. This avoids ambiguities when
implementing the interface to a PHR.
• We added an AbstractSourcesProvider class as an
interface for retrieving data from patient health
records. It defines two methods: one to check
whether the data for a given source can be re-
trieved, and one for actually retrieving the data.
Multiple derived classes (SourcesProvider) can be
implemented, for example for accessing different
databases. They are registered at an instance of
a SourcesProviderManager class, which iterates
over all SourcesProvider instances and queries
whether they can provide a given source and re-
trieve its data. To facilitate the implementation of
a specific SourcesProvider for a guideline model,
a description of all data items is exported as a
JSON file. This description includes the data item
names, data types, and codes from terminology
systems. It can be used by developers who imple-
ment the interface to a PHR as a reference, to pre-
vent the need to use our graphical editor to lookup
the data items.
• Furthermore, our implementation includes a con-
cept for translations. In expressions like task cap-
tions and descriptions, a function tr() can be used
to translate strings:
tr(”text”) → ”text”
translated
(1)
The input string is looked up in a translation map-
ping and the translation is returned. It can be con-
figured which translation mapping shall be used
by the guideline engine, thus it is possible to trans-
late the same guideline into multiple languages.
2.1.1 Graphical Editor
We developed a graphical editor to provide a user-
friendly authoring environment. It follows a low-code
approach to enable users with little programming ex-
pertise to develop CIG models. There are similar vi-
sual programming tools for PROforma in the Tallis
and Alium software suites, but they use the PROforma
syntax to store the guideline models, which we do not
support in favor of RDF.
Our editor is based on the rapid prototyping plat-
form MeVisLab (?), which has a graphical editor for
A Python SDK for Authoring and using Computer-interpretable Guidelines
101

developing networks of image processing and visual-
ization modules. We adapted the graphical editor to
allow for creating task networks of our CIG.
Guideline models are stored as MDL documents,
a MeVisLab specific format, which includes informa-
tion that is only needed by the editor, for example, the
2D positions of the tasks. For the use in our guide-
line engine, the guideline models are exported as RDF
documents. Figure ?? and figure ?? show screenshots
of our editor.
Figure 2: Chronic cough guideline model based on a Tallis
example.
Figure 3: User interface with task parameterization, expres-
sion editor, and bookmark viewer.
Further features of the editor are:
a. a bookmark system for storing URLs and PDF files
for quickly viewing web pages and guideline doc-
uments in an integrated viewer,
b. an integrated tester that uses the engine to execute
the guideline models,
c. a graphical user interfaces for task parameteriza-
tion and for defining data items and predicates, and
d. to collect all occurrences of the tr()-function and
generate a Qt Linguist (?) translation source file
(*.ts), which can be edited by translators using Qt
Linguist to generate the translation mapping file
that our engine can load.
3 RESULTS
With our guideline editor at hand, we authored a
guideline model from our CIG. Then we developed
a decision support system, which uses a web applica-
tion to provide the user interface and a Python-based
REST API service to control the guideline engine.
3.1 Authoring the Guideline Model
Our guideline model is based on the German CPG
“S3-Leitlinie Colitis ulcerosa” (?). Authoring it re-
quired some practicing, because it is not only nec-
essary to understand the PROforma components and
tasks, but also how the execution of the engine works
and how the guideline engine interacts with a DSS.
3.1.1 Transition of Task States
A CIG author needs to understand the transition logic
of the task states, which are: dormant, discarded,
in progress, and completed (?, p. 438). In order to
author a CIG model, tasks are connected in the graph-
ical editor to define which tasks are antecedent tasks
of others. This relation is relevant for the transition of
task states.
When the execution is startet, the engine executes
the guideline model in turns. Each turn, it reviews all
tasks and it checks if the state of a task has to change.
Initially, tasks are dormant. A task stays dormant un-
less it has no antecedent tasks or all of them are com-
pleted or discarded. If all antecedent tasks are dis-
carded, the task is also discarded. If one or more an-
tecedent tasks are completed, the tasks wait condition
is evaluated. If the wait condition is false, then the
task stays dormant. If it is true or if there is no wait
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
102

condition, then the precondition of the task is evalu-
ated. If it exists and evaluates to false, then the task is
discarded, otherwise its state is set to in progress.
After every engine turnaround our DSS checks
which tasks are in progress. If any of these tasks re-
quire user input, which can be either data that cannot
be retrieved from a database or the confirmation of a
task, then our DSS displays a user interface that al-
lows entering data and confirming the task. If the user
provided the input, the DSS forwards it to the engine,
which then validates the input and completes the task,
if the input is valid.
3.1.2 Types of Tasks
PROforma defines different task types: Enquiry, Ac-
tion, Decision, Plan (?; ?). The purpose of these tasks
needs to be understood, as well as a how a DSS that
integrates the guideline engine handles them.
Enquiries are used to access data at a certain en-
gine state. They request sources when they are started
to trigger the retrieval of data for their data items. Ei-
ther the source data is queried automatically through
our SourcesProviderManager instance, or it needs to
be provided by the DSS that integrates the CIG, for
example by asking the user with a dialog. For such an
interactive usage of the CIG, it is relevant where and
when an enquiry is started in the guideline model. For
example, the interactive usability depends on
• how often a user is asked to input data and
• how much data a user has to provide.
The design of the guideline model defines at which
time of execution which data is required. A DSS that
integrates the CIG cannot even this out to improve the
usability. For example, defining a single enquiry that
requests all sources in the beginning of the execution
is possible, but the user might have to input many data
items that will not be used along the executed guide-
line path.
Actions are tasks with the intent to invoke a proce-
dure, which can be an automatic function of the DSS
or an instruction that is displayed to the user. While
authoring a CIG, the connections in the task network,
preconditions, and procedure description are the most
important sub components of actions that need to be
defined. The effect of an action task has to be imple-
mented in the DSS that integrates the guideline en-
gine.
Decisions are tasks that can get complex. They
contain an arbitrary number of decision candidates,
where each candidate can have an arbitrary number
of arguments. Arguments are either in favor of or
against the candidate. They refer to predicates, which
are expressions that can access data items. If the pred-
icate expression evaluates to true, then the argument
applies and it is considered when arguments of the
candidate are weighed up. If the predicate is false,
then the argument does not apply and it is ignored.
For example, an argument against a candidate for a
specific drug therapy might be the allergy to an active
ingredient. If the patient does not have this allergy,
the predicate will evaluate to false and the argument
does not apply.
Arguments in favor of a candidate inrease its net-
support value, arguments against it decrease the net-
support value. In other words: a candidate with a
higher netsupport value than another candidate has
a stronger support based on their arguments. This
information can be either displayed to a user of the
DSS when the decision must be made manually, or the
guideline engine will automatically choose the candi-
date with the highest netsupport value. In addition to
the netsupport value, a candidate can also include an
expression that evaluates to a truth value to specify if
it is recommended.
Plans are tasks that act as structural elements and
are a kind of modularization. They contain an in-
ner task network. In early authoring stages of a CIG
model, when the model often changes, it may help
to omit plans. Otherwise it may be frequently nec-
essary to move tasks in and out of inner task net-
works. Generic tasks, which only include common
task properties like the precondition, can be used as an
antecedent task to serve to some degree as a replace-
ment for plan features like sharing a precondition for
a task network.
3.1.3 Data Items
A work intensive area is the management of data
items. A complex CIG model can require hundreds
of them. For each data item, a type, value range, cap-
tion, description, and code from a terminology system
need to be provided. During authoring it can happen
that certain already modelled data items need to be
discarded or that they have to be replaced by multiple
data items.
3.1.4 CIG Model Tests
As our CIG model grew, we found that we could
break parts of it while continuing with its develop-
ment. Similar to complex software, a complex CIG
model cannot be entirely overlooked. We started to
develop Python unit tests that execute the CIG model
with the CIG engine. The tests provide specific in-
put data and verify that expected execution paths were
encountered. These tests helped us to detect and fix
A Python SDK for Authoring and using Computer-interpretable Guidelines
103

problems early, which is more efficient and effective
than discovering and fixing the problems at a later
stage of the guideline model development.
3.2 Our Decision Support System
The decision support system that we developed in-
cludes a Python based REST API service. It embeds
and runs our guideline engine and allows for load-
ing externally provided guideline models and transla-
tion mapping files. It also includes a SourcesProvider
class that implements the AbstractSourcesProvider
interface and supports querying our database that con-
tains the patient data. An instance of this class is reg-
istered at the SourcesProviderManager instance.
The user interface of our DSS is implemented as a
web application using the Angular framework (?). It
includes our guideline model and sends it to the REST
API service, which then initializes our guideline en-
gine. It further communicates with our REST API
service to execute the guideline.
After each engine turnaround, the REST API ser-
vice queries the engine for all tasks with the state
in progress. If any of these tasks have sources with
requested data items where the data could not be re-
trieved from our database, or if the task needs to be
confirmed by a user, then it returns a JSON object
to the web application. This JSON response contains
the description of the task and its sources. If no task
needs data or requires confirmation, then the next en-
gine turnaround is automatically triggered. This pro-
cedure is repeated until all tasks are either completed
or discarded.
To facilitate user interactions, our web application
implements generic input components for each task
type. All of them display the task caption and de-
scription.
The input component for Actions additionally dis-
plays the procedure text. We have not implemented
any automated triggers for actions.
Another input component exists for tasks that in-
clude sources, which can be enquiries and decisions.
This input component displays the data item values of
the sources and allows entering values for data items
that do not have a value.
For the more complex decisions, there is an ad-
ditional input component to display their candidates
in a vertical list. The candidates are ordered by net-
support value and priority, and information about the
arguments for and against them is added to an expand-
able section, which is collapsed by default. The can-
didates are selectable by the user, and the task will not
complete until a candidate was selected.
Plans have no input component. Instead their in-
ner tasks will be handled when they are in progress.
4 DISCUSSION AND
CONCLUSIONS
The presented decision support system is based on
our Python SDK and allows for determining and pre-
senting clinical practice guideline recommendations
for individual patients. We authored a CIG model
of the “S3-Leitlinie Colitis ulcerosa” (?) guideline.
We did not evaluate it clinically, because the focus of
our work was the realization of the technical concepts.
The model and the engine for CIGs can easily be in-
tegrated into any Python-compatible application such
as decision support systems that will become more
and more relevant when digitization of hospitals in-
creases. In combination with the graphical editor for
the transformation of CPGs to CIGs, arbitrary guide-
lines could be modeled and integrated in decision sup-
port systems.
Besides this we also implemented new function-
ality compared to existing methods. For example,
we added predicates to the PROforma concepts that
allow for a modular use of expressions, so that they
can be reused in other expressions and decision
candidate arguments. This avoids the necessity of
duplicating expressions, which also improves the
maintainability when expressions need to be adapted
throughout the modelling phase of the CIG.
Learnings regarding the development of such systems
are as follows:
• The transformation of a CPG into a CIG re-
quires the mental work of mapping statements and
recommendations to tasks and decision-making
based concepts. This can be challenging and re-
quires practice, if CPGs do not focus on decision-
making or do not describe a process. Some CPGs
rather provide an enumeration of statements and
recommendations.
• Other difficulties are that CPGs are sometimes
ambiguous, vague (?), and target clinical prac-
titioners, who can resolve ambiguities and fill
in missing knowledge using their expert back-
ground. For computer scientists, this means that
they cannot author a CIG on their own, they must
be counseled by clinical experts.
• New CIG authors may be unsure about when to
use an enquiry or a decision. For example, if a
DSS shall allow a user to select a therapy with
a specific drug. It is possible to ask if this drug
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
104

should be used for the treatment using an enquiry
and a boolean data item. But clinical evidence
may describe conditions under which a drug can
be applied as well as alternative drugs. Instead
of an enquiry, a decision should be used. The
drug and its alternatives are modelled as candi-
dates and the conditions are represented by argu-
ments. An argument in favor of a drug could be a
certain indicator, an argument against it could be
an allergy.
• Trustworthiness of CIGs: CPGs need to be trust-
worthy in order to be accepted by clinicians (?).
For example, conflicts of interest of the authors or
different expert opinions about clinical evidence
can impact the CPG trustworthiness, which needs
to be addressed. CIGs that are created on top of
the CPG knowledge inherit these issues. They
may add deviations and errors that occur during
their modelling. This could make it more difficult
to trust CIGs, because digital models are exact,
so they have to be correct, otherwise they could
present false information. But as with other soft-
ware, it is impossible to prove that they are cor-
rect, so testing and reviewing will be necessary
measures to make them trustworthy.
• The recommendations of the guideline “S3-
Leitlinie Colitis ulcerosa” include evidence
grades, recommendation grades, and consensus
grades. PROforma has no direct representation
concepts for these classifications. There could be
situations where they can be mapped to decision
candidate priorities or netsupport values, but that
can also be wrong. For example, prioritizing a
drug therapy because of higher grades can be clin-
ically wrong. A problem is also that a CPG rec-
ommendation may be represented through multi-
ple CIG components. And CIG Components can
also be part of the representation of multiple CPG
recommendations. This makes it difficult to de-
fine the entities in the CIG that should represent
the grades.
In the future, we would like to combine CIGs with lo-
cal decision trees of specific hospitals or sites, maybe
enhanced by deep learning results, to allow for inte-
grative clinical decision support. The integration of
multiple local decision trees into CIGs and the mod-
elling of uncertainty from CPGs into CIGs are impor-
tant steps for future developments.
ACKNOWLEDGEMENTS
This research was funded by the MED²ICIN (Medi-
cal data driving an integrated cost-intelligent model)
project under the Lighthouse Project program of the
Fraunhofer Society.
REFERENCES
Bilici, E., Despotou, G., and Arvanitis, T. N. (2018).
The use of computer-interpretable clinical guidelines
to manage care complexities of patients with multi-
morbid conditions: A review. DIGITAL HEALTH,
4:205520761880492.
Boxwala, A. A., Peleg, M., Tu, S., Ogunyemi, O., Zeng,
Q. T., Wang, D., Patel, V. L., Greenes, R. A., and
Shortliffe, E. H. (2004). GLIF3: a representation for-
mat for sharable computer-interpretable clinical prac-
tice guidelines. Journal of Biomedical Informatics,
37(3):147–161.
Bury, J., Fox, J., and Sutton, D. (2001). The PRO-
forma guideline specification language: Progress and
prospects. Studies in Health Technology and Infor-
matics, 83:13–29.
Bury, J., Hurt, C., Roy, A., Cheesman, L., Bradburn, M.,
Cross, S., Fox, J., and Saha, V. (2005). LISA: a web-
based decision-support system for trial management
of childhood acute lymphoblastic leukaemia. British
Journal of Haematology, 129(6):746–754.
Cancer Research UK (2021).
Tallis: Chronic cough example.
http://archive.cossac.org/tallis/Sample03 Chronic
cough.htm. Accessed: 2021-09-08.
Committee on Standards for Developing Trustworthy Clin-
ical Practice Guidelines, Board on Health Care Ser-
vices, Institute of Medicine, Graham, R., Mancher,
M., Wolman, D. M., Greenfield, S., and Steinberg, E.,
editors (2011). Clinical Practice Guidelines We Can
Trust. National Academies Press (US). Pages: 13058.
Coulson, A. S., Glasspool, D. W., Fox, J., and Emery,
J. (2001). RAGs: A novel approach to comput-
erized genetic risk assessment and decision support
from pedigrees. Methods of Information in Medicine,
40(4):315–322.
de Clercq, P. A., Blom, J. A., Korsten, H. H., and Has-
man, A. (2004). Approaches for creating computer-
interpretable guidelines that facilitate decision sup-
port. Artificial Intelligence in Medicine, 31(1):1–27.
Emery, J., Walton, R., Murphy, M., Austoker, J., Yudkin, P.,
Chapman, C., Coulson, A., Glasspool, D., and Fox,
J. (2000). Computer support for interpreting family
histories of breast and ovarian cancer in primary care:
Comparative study with simulated cases. British Med-
ical Journal, 321(7252):28–32.
Fox, J. (2017). Cognitive systems at the point of care: The
CREDO program. Journal of Biomedical Informatics,
68:83–95.
A Python SDK for Authoring and using Computer-interpretable Guidelines
105

Fox, J., Johns, N., Lyons, C., Rahmanzadeh, A., Thom-
son, R., and Wilson, P. (1997). PROforma: a gen-
eral technology for clinical decision support systems.
Computer Methods and Programs in Biomedicine,
54(1):59–67.
Google LLC and the Angular community (2021). An-
gular - the modern web developer’s platform.
https://angular.io/. Accessed: 2021-09-10.
Heckel, F., Schwier, M., and Peitgen, H.-O. (2009). Object-
oriented application development with mevislab and
python. In Informatik 2009 – Im Focus das Leben,
pages 1338–1351. Gesellschaft f
¨
ur Informatik e. V.
HL7 (2021a). Fhir: standard for health care data exchange.
https://www.hl7.org/fhir/. Accessed: 2021-09-08.
HL7 (2021b). Fhir: standard for health care data exchange.
https://www.hl7.org/fhir/datatypes.html#Coding. Ac-
cessed: 2021-09-08.
Khodambashi, S., Slaughter, L., and Nytrø, Ø. (2015).
Computer-interpretable clinical guidelines: A review
and analysis of evaluation criteria for authoring meth-
ods. In Studies in health technology and informatics,
volume 216.
Kogan, A., Tu, S. W., and Peleg, M. (2018). Goal-driven
management of interacting clinical guidelines for mul-
timorbidity patients. AMIA ... Annual Symposium pro-
ceedings. AMIA Symposium, 2018:690–699.
Kosara, R. and Miksch, S. (2001). Metaphors of movement:
a visualization and user interface for time-oriented,
skeletal plans. Artificial Intelligence in Medicine,
22(2):111–131.
Kucharzik, T., Dignass, A. U., Atreya, R., Bokemeyer,
B., Esters, P., Herrlinger, K., Kannengießer, K.,
Kienle, P., Langhorst, J., L
¨
ugering, A., Schreiber,
S., Stallmach, A., Stein, J., Sturm, A., Te-
ich, N., and Siegmund, B. (2021). Aktual-
isierte S3-Leitlinie Colitis ulcerosa – Living Guide-
line. https://www.awmf.org/leitlinien/detail/ll/021-
009LG.html. Accessed: 2021-09-08.
Lozano, E., Marcos, M., Mart
´
ınez-Salvador, B., Alonso, A.,
and Alonso, J. R. (2010). Experiences in the devel-
opment of electronic care plans for the management
of comorbidities. In Knowledge Representation for
Health-Care. Data, Processes and Guidelines, vol-
ume 5943, pages 113–123. Springer Berlin Heidel-
berg. Series Title: Lecture Notes in Computer Sci-
ence.
Miles, A., Chronakis, I., Fox, J., and Mayer, A. (2017). Use
of a computerised decision aid (DA) to inform the de-
cision process on adjuvant chemotherapy in patients
with stage II colorectal cancer: development and pre-
liminary evaluation. BMJ open, 7(3):e012935.
Patkar, V., Acosta, D., Davidson, T., Jones, A., Fox, J., and
Keshtgar, M. (2012). Using computerised decision
support to improve compliance of cancer multidisci-
plinary meetings with evidence-based guidance. BMJ
Open, 2(3):e000439.
Patkar, V., Hurt, C., Steele, R., Love, S., Purushotham,
A., Williams, M., Thomson, R., and Fox, J. (2006).
Evidence-based guidelines and decision support ser-
vices: a discussion and evaluation in triple assessment
of suspected breast cancer. British Journal of Cancer,
95(11):1490–1496.
Peleg, M. (2013). Computer-interpretable clinical guide-
lines: A methodological review. Journal of Biomedi-
cal Informatics, 46(4):744–763.
Peleg, M., Fox, J., Patkar, V., Glasspool, D., Chronakis, I.,
South, M., Nassar, S., Gaglia, J., Gharib, H., Papini,
E., Paschke, R., Duick, D., Valcavi, R., Heged
¨
us, L.,
and Garber, J. (2013). A computer-interpretable ver-
sion of the AACE, AME, ETA medical guidelines for
clinical practice for the diagnosis and management of
thyroid nodules. Endocrine practice : official jour-
nal of the American College of Endocrinology and the
American Association of Clinical Endocrinologists,
20:1–33.
Peleg, M., Tu, S., Bury, J., Ciccarese, P., Fox, J., Greenes,
R. A., Hall, R., Johnson, P. D., Jones, N., Kumar,
A., Miksch, S., Quaglini, S., Seyfang, A., Short-
liffe, E. H., and Stefanelli, M. (2003). Compar-
ing computer-interpretable guideline models: A case-
study approach. Journal of the American Medical In-
formatics Association, 10(1):52–68.
Sonnenberg, F. A. and Hagerty, C. G. (2006). Computer-
interpretable clinical practice guidelines: Where are
we and where are we going? Yearbook of Medical
Informatics, 15(1):145–158.
Stanford University (2021). Prot
´
eg
´
e.
https://protege.stanford.edu/products.php. Accessed:
2021-09-08.
Stan
ˇ
cin, I. and Jovi
´
c, A. (2019). An overview and compar-
ison of free python libraries for data mining and big
data analysis. In 2019 42nd International Convention
on Information and Communication Technology, Elec-
tronics and Microelectronics (MIPRO), pages 977–
982.
Sutton, D. R. and Fox, J. (2003). The syntax and semantics
of the PRO forma guideline modeling language. Jour-
nal of the American Medical Informatics Association,
10(5):433–443.
The Qt Company (2021). Qt linguist manual.
https://doc.qt.io/qt-5/qtlinguist-index.html. Ac-
cessed: 2021-09-09.
Tural, C., Ruiz, L., Holtzer, C., Schapiro, J., Viciana, P.,
Gonz
´
alez, J., Domingo, P., Boucher, C., Rey-Joly, C.,
Clotet, B., and Havana Study Group (2002). Clinical
utility of HIV-1 genotyping and expert advice: the ha-
vana trial. AIDS (London, England), 16(2):209–218.
W3C (2021a). Resource description framework (rdf).
https://www.w3.org/RDF/. Accessed: 2021-09-08.
W3C (2021b). Turtle syntax.
https://www.w3.org/TR/turtle/. Accessed: 2021-
09-08.
Walton, R., Gierl, C., Yudkin, P., Mistry, H., Vessey, M.,
and Fox, J. (1997). Evaluation of computer support for
prescribing (CAPSULE) using simulated cases. BMJ
(Clinical research ed.), 315:791–5.
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
106
