
GAMS: Graph Augmentation with Module Swapping
Alessandro Bicciato and Andrea Torsello
Department of Environmental Sciences, Informatics and Statistics, Ca’ Foscari University of Venice, Italy
Keywords:
Augmentation, Motif, Swapping.
Abstract:
Data augmentation is a widely adopted approach to solve the large-data requirements of modern deep learning
techniques by generating new data instances from an existing dataset. While there is a huge literature and
experience on augmentation for vectorial or image-based data, there is relatively little work on graph-based
representations. This is largely due to complex, non-Euclidean structure of graphs, which limits our abilities
to determine operations that do not modify the original semantic grouping.
In this paper, we propose an alternative method for enlarging the graph set of graph neural network datasets by
creating new graphs and keeping the properties of the originals. The proposal starts from the assumptions that
the graphs compose a set of smaller motifs into larger structures. To this end, we extract modules by grouping
nodes in an unsupervised way, and then swap similar modules between different graphs reconstructing the
missing connectivity based on the original edge statistics and node similarity. We then test the performance
of the proposed augmentation approach against state-of-the-art approaches, showing that on datasets, where
the information is dominated by structure rather than node labels, we obtain a significant improvement with
respect to alternatives.
1 INTRODUCTION
Deep Learning techniques have proved to be effective
in tackling a wide range of real-world tasks due to
their ability to effectively learn low- to medium-level
representation for the problems at hand, and to pro-
vide more complex decision boundaries to reflect the
complexity of the problems. However, this results in
generally much larger hypothesis spaces and a greater
tendency to over-fitting what in general needs to be
balanced by means of much larger amounts of train-
ing data. One approach to make up for the large data
requirements of these approaches is data augmenta-
tion, i.e. the generation of new plausible that simu-
lates a re-sampling of the space of possible training
instances, by transforming and recombining the exist-
ing data. Augmentation operations are easily devised
for computer vision and natural language process-
ing tasks where one can express nuisance data trans-
formations that modify the input without modifying
the original semantic grouping. For example, object
translation and rotation, occlusion, and background
substitutions can readily be used on object detection
tasks to increase the number of training images. For
more general vectorial data nuisance, transformations
can be harder to define, but one can use continuity
assumptions to artificially increase the sample den-
sity of the feature space. However, even continuity
assumptions have to be given up when dealing with
graphs-based representations.
Graphs have long been used as a powerful abstrac-
tion for a wide variety of real-world data where struc-
ture plays a key role, from collaborations (Lima et al.,
2014) to biological data (Gilmer et al., 2017; Ye et al.,
2015). While Graph Neural Networks (GNNs) have
gained increasing traction in the machine learning
community for being able to effectively tackle clas-
sification problem abstracted in terms of graphs, the
increased complexity of deep models expresses itself
in these approaches as well, resulting in a tendency to
over-fitting and beyond what can be addressed with
simple dropout techniques, to the point that represen-
tations of nodes belonging to different classes become
indistinguishable when stacking multiple layers, seri-
ously hurting the model accuracy (Chen et al., 2019).
In general, this leads to a requirement for large dataset
that can be harder to come by than in the case of im-
ages and text, since graph-based representation reside
at a higher semantic level and cannot be acquired as
easily.
However, data augmentation techniques for
graphs is still under-researched, with most methods
performing simple node and edge edit operations
with little consideration of the larger structure of the
Bicciato, A. and Torsello, A.
GAMS: Graph Augmentation with Module Swapping.
DOI: 10.5220/0010822400003122
In Proceedings of the 11th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2022), pages 249-255
ISBN: 978-989-758-549-4; ISSN: 2184-4313
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
249

graphs. In this paper we propose a novel augmenta-
tion approach called Graph Augmentation with Mod-
ule Swapping (GAMS) which tries to automatically
detect coherent portions of the graphs motifs and aug-
ment the dataset producing new graphs by swapping
similar motifs between different graphs. The idea be-
hind this approach is that the structural representa-
tions do satisfy some form of compositionality rule
just like image and text data, and it is formed by
the composition of several simpler coherent substruc-
tures loosely connected with one another according to
some unknown compositionality rule. Under this as-
sumption, swapping the modules is equivalent to in-
terchanging base substructures into existing composi-
tion templates.
2 STATE OF THE ART
Approaches for augmenting graph data are relatively
limited and mostly consist in heuristics to select
which elements of the structure (nodes/edges) to per-
turb in order to generate a new graph from a sin-
gle sample. One of the simplest and most used ap-
proaches for graph augmentation is graph perturba-
tion, which consists in a series of edge addition or
removal operations between existing nodes of a sin-
gle graph. This presents several degrees of freedom
and a large number of parameters. In particular, edge
selection can be random or follow a given heuristic,
making this more of a meta-approach with several dif-
ferent instances depending on the perturbation strat-
egy adopted. Perturbation approaches are in general
simple to implement and fast in their execution. How-
ever, in their simplest incarnation they give little to no
control over what part of the structure gets modified,
resulting in graphs that have very little to do with the
originals, and which do not maintain the underlying
semantic label. For instance, the algorithm might add
several edges in a sparse area of the graph or remove
them from a dense area resulting in graphs that are
very dissimilar from the original.
The simplest instance of edge perturbation is
given by DropEdge (Rong et al., 2019), which con-
sists in randomly dropping edges of the input graph
for each training iteration. This method was designed
to alleviate over-fitting and over-smoothing when
training Graph Convolutional Networks (GCNs).
Through DropEdge, we are actually generating differ-
ent randomly deformed copies of the original graph,
thus increasing randomness and diversity of the in-
put data, reducing the risk of over-fitting. Second,
DropEdge can also be treated as a message passing
reducer. In GCNs, the message passing between adja-
cent nodes is conducted along edge paths. Removing
certain edges renders node connections sparser, thus
avoiding over-smoothing to some extent when GCN
goes very deep (Rong et al., 2019). This algorithm is
well suited for deep learning algorithms, with DropE-
dge being executed at the end of each training epoch,
generating the new dataset for the next epoch.
The AdaEdge algorithm (Chen et al., 2019) op-
timizes the graph topology based on the model pre-
dictions. The method consists in iteratively training
GNN models and conduct edge remove/add opera-
tions based on the prediction to adaptively adjust the
graph for the learning target. Experimental results
in general cases show that this method can signifi-
cantly relieve the over-smoothing issue and improve
model performance, which further provides a com-
pelling perspective towards better GNNs performance
(Chen et al., 2019). More specifically, GNNs are
trained on the original graph and then the graph topol-
ogy is adjusted based on the prediction result of the
model by deleting inter-class edges and adding intra-
class edges. The GNN is then retrained on the updated
graph, and the topology optimization and model re-
training are iterated multiple times.
The GAUG (Zhao et al., 2020) methods follow a
similar concept to DropEdge and AdaEdge, driving
the augmentation based on the results of the trained
classifier. The goal is to improve node classifica-
tion by mitigating propagation of noisy edges. Neural
edge predictors like GAE (Kipf and Welling, 2016)
are able to latently learn class-homophilic tenden-
cies in existent edges that are improbable, and nonex-
istent edges that are probable (Zhao et al., 2020).
GAUG key idea is to leverage information inherent in
the graph to predict which non-existent edges should
likely exist, and which existent edges should likely be
removed in the graph G to produce modified graph(s)
G
m
to improve model performance.
• GAUG-M: this procedure starts using an edge pre-
dictor function to obtain edge probabilities for all
possible and existing edges in G. The role of the
edge predictor is flexible and can generally be re-
placed with any suitable method. Then we use
the predicted edge probabilities, we deterministi-
cally add (remove) new (existing) edges to create
a modified graph G
m
, which is used as input to a
GNN node-classifier.
This method is tough for modified-graph setting,
i.e. when we apply one or multiple graph trans-
formation operation f : G → G
m
, such that G
m
re-
places G for both training and inference.
• GAUG-O: it is complementary to GAUG-M be-
cause it is applied for original-graph setting, i.e.
when we apply many transformations f
i
: G → G
i
m
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
250

C
1
C
2
C
3
C'
1
0.8 0.6 0.4
C'
2
0.5
0.9 0.3
C'
3
0.6 0.7 0.5
C
1
C
2
C
3
C'
1
C'
2
C'
3
G
1
G
2
C
2
n
0
↔ C'
2
n'
2
C
2
n
1
↔ C'
2
n'
1
↔ C'
2
n'
0
n
0
n
1
C
2
n'
0
n'
1
n'
2
C'
2
n
0
n
1
n'
1
n'
2
n'
0
n'
0
n'
1
n'
2
n
0
n
1
C
2
C
1
C
3
C'
1
C'
3
C'
2
Take 2 graphs from the
dataset
Divide in clusters
Compare the clusters and
select the most similar
HKS on the nodes of the
selected clusters
Associate the nodes of one
cluster to the ones of the other
Build new graphs
Figure 1: Schematic representation of the algorithm.
for i = 1...N, such that G ∪ {G
i
m
}
N
i=1
may be used
in training, but only G is used for inference.
The method of computation reminisces of the two
steps of GAUG-M and it also uses an edge pre-
diction module for the benefit of node classifica-
tion and aims to improve model generalization.
It does not require discrete specification of edges
to add/remove, is end-to-end trainable, and uti-
lizes both edge prediction and node-classification
losses to iteratively improve augmentation capac-
ity of the edge predictor and classification capac-
ity of the node classifier GNN.
Note that both AdaEdge and GAUG are designed
for node classifications scenarios, where the whole
dataset consists of a single graph, and the classifier
produces a label per node. This way the classifica-
tion response can be localized in the structure and can
be used to drive the edit operations. However, in a
graph classification scenario, where the dataset con-
sists of several graph and the task is to classify the
graph themselves, the label cannot be as directly used
to drive the augmentation.
GraphCL (You et al., 2021) is a recent augmen-
tation technique mainly used for learning unsuper-
vised, semi-supervised and supervised representation
of graph data, for both node classification and graph
classification tasks. The approach is based on a graph
contrastive learning (GraphCL) framework that can
combine four different augmentation methods: node
dropping, edge perturbation, attribute masking and
subgraph.
• Node Dropping: Given the graph G, node drop-
ping randomly discards a specified proportion of
vertices along with their connections. The under-
lying idea behind this is that ”occluding” a small
portion of vertices does not affect the semantic la-
beling G;
• Edge Perturbation: perturbs the connectivities in
G by randomly adding or dropping a given ratio of
edges. The underlying idea is that that the seman-
tic label of G has a level of robustness to variation
in the edge connectivity patterns;
• Attribute Masking: attribute masking eliminates
some vertex attributes, forcing the model to re-
cover them using contextual information. The
underlying assumption is that model predictions
should be robust with respect to missing partial
vertex attributes.
• Subgraph: This method samples a subgraph from
G using random walk. It assumes that the seman-
tics of G is preserved in its (partial) local struc-
ture.
The purpose of the GraphCL framework is to produce
graph representations of similar or better generaliz-
ability, transferability, and robustness compared to the
competing approaches.
3 CLUSTERING
AUGMENTATION
Our proposed graph augmentation algorithm is de-
signed for a graph classification task. The idea be-
hind the proposal is that, just like images and text,
graph representations are obtained through the com-
position of smaller structural components, motifs, that
are in general internally more coherent than the rest of
the structure. The goal of a deep classifier is then to
implicitly capturing both these motifs and their com-
positional rules. In our proposal, with a schematic
representation in Figure 1, we extract the motifs in
an unsupervised way through a graph-clustering ap-
proach, thus we partition each graph in the dataset
into a fixed number of clusters C. Let G
a
= (V
a
,E
a
)
and G
b
= (V
b
,E
b
) be two graphs belonging to the
same semantic class, with V
a
and V
b
being the node
sets, and E
a
⊆ V
a
×V
a
and E
b
⊆ V
b
×V
b
the edge sets
of the two graph. Then we find the pair of motifs, one
from each graph, that are most similar to one another,
and swap them rebuilding the connections with the
rest of the graphs’ nodes. This gives us two new per-
turbed graphs G
0
a
and G
0
b
to be added to the training
set.
More generally, given a set D
`
of graphs having
GAMS: Graph Augmentation with Module Swapping
251

Table 1: Overview of the datasets tested.
Dataset # Graphs Classes Avg. Nodes Avg. Edges Avg. Degree
DD 1178 2 284.32 715.66 2.517
MUTAG 188 2 17.93 19.79 1.103
PROTEINS 1113 2 39.06 72.82 1.864
ENZYMES 600 6 32.63 62.14 1.904
MSRC 21 563 20 77.52 198.32 2.558
NCI1 4110 2 29.87 32.30 1.081
IMDB-MULTI 1500 3 13.00 65.94 5.072
COLLAB 5000 3 74.49 2457.78 32.994
REDDIT-BINARY 2000 2 429.63 497.75 1.158
the same label `, we can partition the graph in the
set, and augment the set by performing high similar-
ity swaps between partition of all the same-labeled
graphs.
The first step of our approach is to partition the
nodes of each graph into clusters. The subgraphs
induced by each partition are candidates for our ex-
tracted motifs. Any suitable clustering approach can
be used for motif extraction, but in our implementa-
tion we used the spectral clustering implementation
provided by the Scikit Learn library, which is a vari-
ation of the normalized cut algorithm (Shi and Malik,
2000).
After the extraction of the motif candidates we
measure the similarity between each pair of motif
using the Weisfeiler-Lehman Kernel (Weisfeiler and
Lehman, 1968). The key idea of the Weisfeiler-
Lehman kernel is to replace the label of each vertex
with a multiset label, consisting of the original label
of the vertex and the sorted set of labels of its neigh-
bors. The resultant multiset is then compressed into
a new, short label. This relabeling procedure is then
repeated for h iterations.
Using the resulting kernel to the second cluster,
we get the transformation, i.e. the degree of similarity
between the pair of clusters.
With the motif similarities to hand, we can per-
form the swap. Given two graph with he same class
label, we select and swap the most similar clusters
from one graph to the other. Once the graph and mo-
tifs to be swapped are selected, we need a way to re-
stitch the motifs to the new graph, i.e. re-create the
connectivity between the swapped-in motif and the
rest of the graph. This is done by taking all the edges
outgoing from the motif and connecting them to the
node in the new graph that is most similar to the node
the edge was originally connected to.
More formally, let G
a
= (V
a
,E
a
) and G
b
=
(V
b
,E
b
) be the two graphs, and assume we decided
to swap motif M
a
= (V
M
a
,E
M
a
) from G
a
with motif
M
b
= (V
M
b
,E
M
b
) with V
M
a
⊆ V
a
, E
M
a
= E
a
∩ V
M
a
×
V
M
a
. Let E
⊥
M
a
and E
⊥
M
b
be the set of edges connecting
M
a
and M
b
with the rest of the respective graphs, i.e.
E
⊥
M
a
= {(u,v) ∈ E
a
|u ∈ M
a
and v 6∈ M
a
} (1)
E
⊥
M
b
= {(u,v) ∈ E
b
ku ∈ M
b
and v 6∈ M
b
}.
Then, to stitch M
a
into G
b
\ M
b
, for every (u,v) ∈
E
M
a
we find the node v
0
∈ V
b
\V
M
b
most similar to v
and create an edge from u to v
0
in the new graph.
The similarity between the nodes in the two struc-
ture can be gauged by any measure incorporating
structural information and/or vertex attributes. In
out experiments we used the Heat Kernel Signa-
ture (H
¨
ormann, 2014) to characterize the structure
around each node and used the cosine similarity to
measure the similarity between the signatures.
Once the similarities between all nodes in V
a
\V
M
a
and V
b
\ V
M
b
are computed, we cast the problem
of matching the outgoing connections in E
M
a
with
V
b
\ V
M
b
as a bipartite matching problem, thus find-
ing the set of nodes in V
b
\V
M
b
to which to connect
the outgoing edges E
⊥
M
a
from motif M
a
. The same is
performed in reverse to stitch M
b
into G
a
\ M
b
. In our
implementation we used the auction algorithm (Bert-
sekas, 1992) to solve the bipartite matching problem.
The code for our approach is available at
https://gitlab.com/ripper346-phd/graph-cluster-
augmentation.
4 RESULTS
To test the effectiveness of the augmentation al-
gorithm we applied a graph convolutional network
(GCN) to several graph classification tasks and tested
the performance after augmenting the training set by
different amount. The model used embeds each node
by performing multiple rounds of message passing;
aggregate node embeddings into a unified graph em-
bedding (readout layer); train a final classifier on the
graph embedding. The readout layers took the aver-
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
252
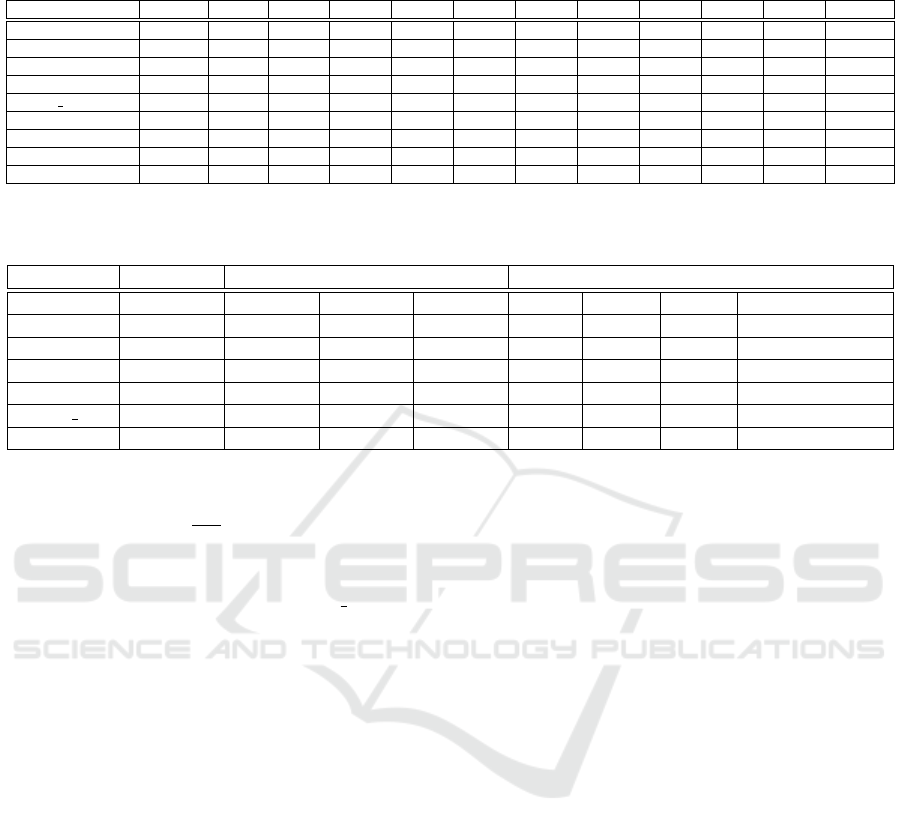
Table 2: Test accuracy of 200th epoch of class classification training with increasing number of graphs. The bold numbers are
the best results.
Datasets Original +50% +100% +200% +300% +400% +500% +600% +700% +800% +900% +1000%
DD 0.6907 0.7288 0.7055 0.6945 0.7878 0.8091 0.7381 0.8176 0.8256 0.7973 0.8141 0.8750
MUTAG 0.7368 0.8421 0.7763 0.7876 0.7616 0.8989 0.8230 0.8561 0.9070 0.8525 0.8298 0.8575
PROTEINS 0.7315 0.6543 0.6898 0.6836 0.7269 0.7185 0.7091 0.7070 0.6950 0.7078 0.7079 0.7378
ENZYMES 0.3500 0.2961 0.2594 0.2821 0.2432 0.2433 0.2640 0.2120 0.2652 0.2815 0.2617 0.2453
MSRC 21 0.8761 0.9231 0.8717 0.9467 0.9246 0.9520 0.9630 0.9556 0.9512 0.9635 0.9565 0.9677
NCI1 0.6910 0.6285 0.6582 0.6215 0.6176 0.6441 0.6341 0.6318 0.6242 0.6381 0.6346 0.6337
IMDB-MULTI 0.3967 0.3756 0.3850 0.3944 0.3975 0.4189 0.4373 0.4728 0.4329 0.4646 0.4485 0.4603
COLLAB 0.7240 0.7113 0.7480 0.7547 0.7733 0.7700 - - - - - - - - - - - - - - - - - -
REDDIT-BINARY 0.7575 0.8033 0.8300 - - - - - - - - - - - - - - - - - - - - - - - - - - -
Table 3: Test accuracy of 200th epoch of class classification training with increasing number of graphs compared to three
different GraphCL runs. The bold numbers are the best results.
Datasets Untouched GraphCL GAMS
random 4 random 3 random 2 +50% +100% +300% best
DD 0.6907 0.5847 0.5339 0.6186 0.7288 0.7055 0.7878 0.8750 (+1000%)
MUTAG 0.7368 0.7368 0.6842 0.5263 0.8421 0.7763 0.7616 0.9070 (+700%)
PROTEINS 0.7315 0.5964 0.5830 0.6457 0.6543 0.6898 0.7269 0.7378 (+1000%)
ENZYMES 0.3500 0.1917 0.2083 0.2667 0.2961 0.2594 0.2432 0.2961 (+50%)
MSRC 21 0.8761 0.6637 0.6814 0.7168 0.9231 0.8717 0.9246 0.9677 (+1000%)
NCI1 0.6910 0.5182 0.4927 0.5049 0.6285 0.6582 0.6176 0.6582 (+100%)
age of node embeddings:
x
G
=
1
|V |
∑
v∈V
x
(L)
v
(2)
We performed the tests on different datasets:
D&D, Mutag, Proteins, Enzymes, MSRC 21, NCI1,
COLLAB, IMDB-Multi and Reddit-Binary. Table 1
provides a brief overview of the datasets used.
In our experiments we divided each graph into
five clusters and then recombined the motifs increas-
ing the size of the training set by increasing amounts,
starting from an increase off 50% all the way to an
increase of 1000% of the original size. Table 2 shows
the results of the classification on the test set using
GCN classifiers trained on the augmented training set
and on the original unaugmented training set. The
best result for each dataset is in bold. From this table
we can see that the augmentation provides an advan-
tage in most datasets, with some datasets exhibiting a
dramatic increase in performance after the augmenta-
tion procedure. It is worth noting that the best results
are for datasets containing bigger and denser graphs,
where the structural component is more determinant
in classification. On the other hand, with datasets
such as ENZYMES and NCI1, which are generally
sparser and with more complex vertex attributes, the
augmentation seems to not provide any advantage.
Also, neither the classification nor the node match-
ing and stitching approaches that we adopted make
use of the vertex attributes, and more attribute-aware
might improve the results on these datasets. Nonethe-
less, it is important to note that for all datasets the
performance has a clear increasing trend as the level
of augmentation increases. This can also be seen in
Figure 2 where we display the classification perfor-
mance at the varying training epochs for various lev-
els of augmentation of the training set. We can see
that large augmentations increase convergence speed
and reduce the volatility of the accuracy throughout
the training process, as well as increase the final per-
formance of the network.
We compared the results obtained with our
augmentation approach with the semi-supervised
GraphCL. In particular we use three runs of GraphCL:
• random4: use the degree feature, one hot degree
feature with max degree 100, a
k3
feature, and
with random selection between node dropping,
edge perturbation, attribute masking and subgraph
methods;
• random3: use the degree feature, one hot degree
feature with max degree 100, a
k3
feature, and with
random selection between node dropping, edge
perturbation and subgraph methods;
• random2: use the degree feature, one hot degree
feature with max degree 100, a
k3
feature, and with
random selection between node dropping and sub-
graph methods.
In Table 3 we can see the results on some of the
datasets used for GAMS. The results of our algorithm
are always better than those obtained with GraphCL
(for clarity in the table we compared only to +50%,
+100%, +300% but we can still take all the results of
GAMS from Table 2), even in those instances where
GAMS: Graph Augmentation with Module Swapping
253
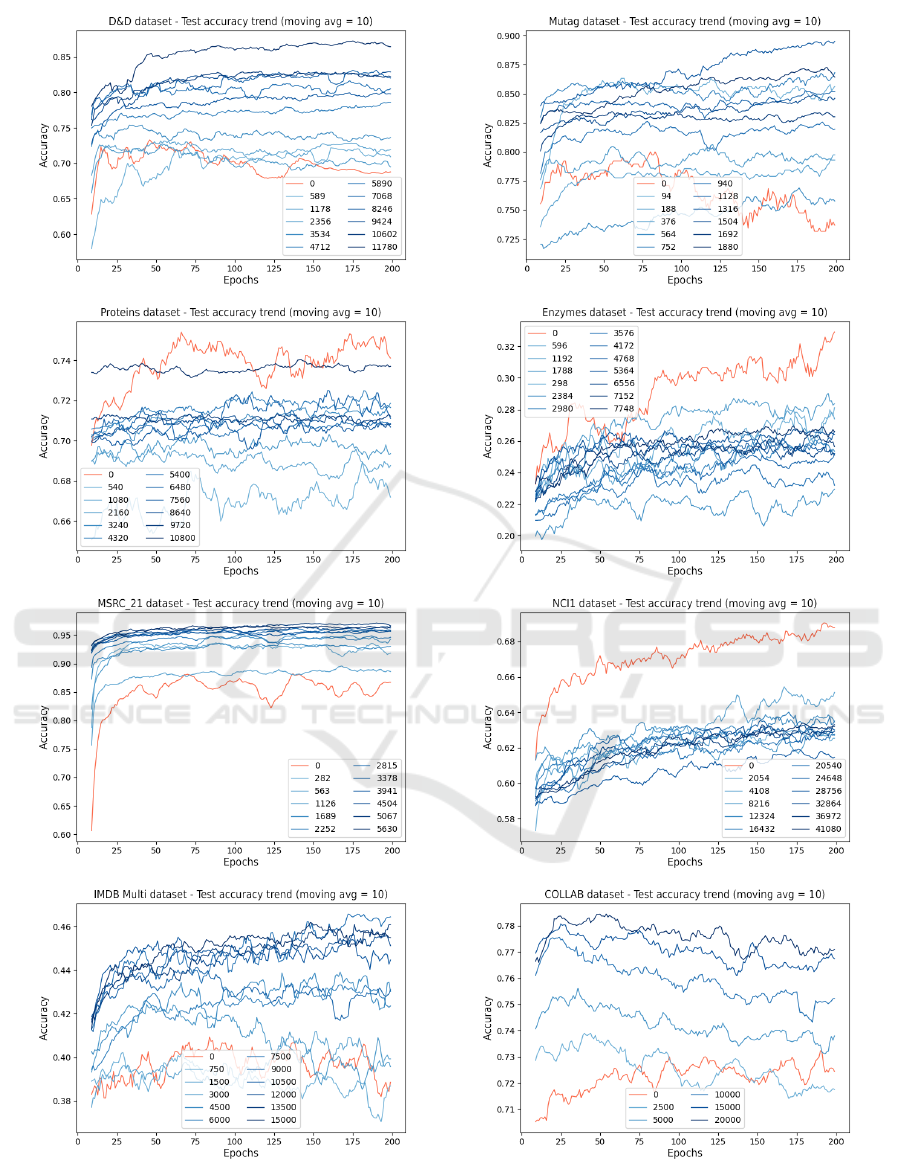
Figure 2: Accuracy results of the test set for the datasets’ graph classification training with 5 cluster division. For clarity it is
shown the moving average of the results with W = 10. The red curve is the untouched dataset training, the other curves are the
augmented runs, where the darker the color the bigger is the augmentation. In the legend are reported the number of graphs
added to the original dataset.
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
254

GAMS decreases the performance with respect to the
original training set, confirming the hypothesis that
in these datasets the structure is not as important for
classification.
5 CONCLUSION
In this paper, we presented a new method for graph
augmentation. This method, works under the assump-
tion that the graphs are formed by composing simpler
cohesive substructures (motifs), and operates by ex-
tracting and swapping these substructures from train-
ing graph with the same label. This provides an aug-
mentation approach that is data-driven, in the sense
that the swapped-in motifs are directly observed from
the dataset, but that not require to be learned nor it
is in any way coupled with the learning algorithm
adopted for the final classification problem.
Our experimental evaluation showed that the ap-
proach is capable of providing very substantive in-
creases in performance for datasets where the struc-
tural information is determinant for classification,
consistently yielding better results than competing ap-
proaches at the state of the art, and in some instances
allowing state-of-the-art classification performances
even with a simple GCN scheme.
REFERENCES
Bertsekas, D. P. (1992). Auction algorithms for network
flow problems: A tutorial introduction. LIDS-P-2108.
Chen, D., Lin, Y., Li, W., Li, P., Zhou, J., and Sun, X.
(2019). Measuring and relieving the over-smoothing
problem for graph neural networks from the topologi-
cal view. arXiv:1909.03211.
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O., and
Dahl, G. E. (2017). Neural message passing for quan-
tum chemistry. In International conference on ma-
chine learning, pages 1263–1272. PMLR.
H
¨
ormann, T. (2014). Heat kernel signature.
Kipf, T. N. and Welling, M. (2016). Variational graph au-
toencoders. arXiv:1611.07308.
Lima, A., Rossi, L., and Musolesi, M. (2014). Coding to-
gether at scale: Github as a collaborative social net-
work. In Eighth international AAAI conference on we-
blogs and social media.
Rong, Y., Huang, W., Xu, T., and Huang, J. (2019). Drope-
dge: Towards deep graph convolutional networks on
node classification. arXiv:1907.10903v4.
Shi, J. and Malik, J. (2000). Normalized cuts and image
segmentation. IEEE Trans. on Pattern Analysis and
Machine Intelligence, 22(8):888–905.
Weisfeiler, B. and Lehman, A. A. (1968). A reduction of
a graph to a canonical form and an algebra arising
during this reduction. Nauchno-Technicheskaya Infor-
matsia, pages 2(9):12–16.
Ye, C., Comin, C. H., Peron, T. K. D., Silva, F. N., Ro-
drigues, F. A., Costa, L. d. F., Torsello, A., and Han-
cock, E. R. (2015). Thermodynamic characterization
of networks using graph polynomials. Physical Re-
view E, 92(3):032810.
You, Y., Chen, T., Sui, Y., Chen, T., Wang, Z., and Shen,
Y. (2021). Graph contrastive learning with augmenta-
tions. arXiv:2010.13902v3.
Zhao, T., Liu, Y., Neves, L., Woodford, O., Jiang, M., and
Shah, N. (2020). Data augmentation for graph neural
networks. arXiv:2006.06830v2.
Zhou, J., Shen, J., Yu, S., Chen, G., and Xuan, Q. (2020).
M-evolve: Structural-mapping-based data augmenta-
tion for graph classification. CoRR, abs/2007.05700.
GAMS: Graph Augmentation with Module Swapping
255
