
Automatic Label Detection in Chest Radiography Images
Jo
˜
ao Pedrosa
1,2 a
, Guilherme Aresta
1,2 b
, Carlos Ferreira
1,2 c
,
Ana Maria Mendonc¸a
1,2 d
and Aur
´
elio Campilho
1,2 e
1
Institute for Systems and Computer Engineering, Technology and Science (INESC TEC), Porto, Portugal
2
Faculty of Engineering of the University of Porto (FEUP), Porto, Portugal
Keywords:
Chest Radiography, Deep Learning, Object Detection, Classification Bias, Markers.
Abstract:
Chest radiography is one of the most ubiquitous medical imaging exams used for the diagnosis and follow-up
of a wide array of pathologies. However, chest radiography analysis is time consuming and often challeng-
ing, even for experts. This has led to the development of numerous automatic solutions for multipathology
detection in chest radiography, particularly after the advent of deep learning. However, the black-box nature
of deep learning solutions together with the inherent class imbalance of medical imaging problems often leads
to weak generalization capabilities, with models learning features based on spurious correlations such as the
aspect and position of laterality, patient position, equipment and hospital markers. In this study, an automatic
method based on a YOLOv3 framework was thus developed for the detection of markers and written labels
in chest radiography images. It is shown that this model successfully detects a large proportion of markers
in chest radiography, even in datasets different from the training source, with a low rate of false positives per
image. As such, this method could be used for performing automatic obscuration of markers in large datasets,
so that more generic and meaningful features can be learned, thus improving classification performance and
robustness.
1 INTRODUCTION
Chest radiography (CXR), also known as chest x-ray,
is one of the most ubiquitous medical imaging ex-
ams and remains extremely advantageous thanks to
its wide availability, low cost, portability and low ra-
diation dosage in comparison to other ionizing imag-
ing modalities. Moreover, radiologists typically use
CXRs for the diagnosis or screening of multiple con-
ditions associated to the chest wall and the lungs as
well as the heart and greater vessels. Nevertheless,
the assessment of CXR images is time consuming and
often challenging, even for experts. Furthermore, the
large quantity of CXR exams acquired per day can
lead to an unmanageable workload for radiologists,
leading to misdiagnosis.
As such, computer-aided diagnosis (CAD) sys-
tems for CXR pathology detection have long been
proposed, providing a valuable 2
nd
opinion for radi-
a
https://orcid.org/0000-0002-7588-8927
b
https://orcid.org/0000-0002-4856-138X
c
https://orcid.org/0000-0001-6754-6495
d
https://orcid.org/0000-0002-4319-738X
e
https://orcid.org/0000-0002-5317-6275
ologists or screening abnormal cases that radiologists
should assess visually. Traditional machine learning
approaches have mostly been applied for the detection
of a specific disease (Qin et al., 2018), specifically,
for lung nodule and tuberculosis detection. However,
these algorithms fail to represent the wide array of
pathologies encountered in the clinical environment.
The recent advent of deep learning, as well as the
release of large CXR datasets such as ChestXRay-8
(Wang et al., 2017) and CheXpert (Irvin et al., 2019),
have fostered the development of multi-disease detec-
tion approaches, while simultaneously improving per-
formance in the detection of single pathologies (Irvin
et al., 2019).
However, the intrinsic nature of deep learning
techniques, where image features are learned from an
image-level label (normal vs pathological or pathol-
ogy A vs B) can lead to unexpected behaviour and
low explainability. In fact, in complex images such
as CXR and in severe imbalance of classes and data,
there is no guarantee that the decisions made by a
deep learning system are representative of a clinical
finding and not a spurious correlation of the data. In-
deed, recent algorithms for COVID-19 detection in
Pedrosa, J., Aresta, G., Ferreira, C., Mendonça, A. and Campilho, A.
Automatic Label Detection in Chest Radiography Images.
DOI: 10.5220/0010888100003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 63-69
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
63

CXR were shown to have learned correlations in the
data rather than clinical information (DeGrave et al.,
2020). Because most models for COVID-19 detec-
tion are trained with a mixture of negative COVID-19
pre-pandemic CXRs and positive COVID-19 cases, it
becomes simpler to learn shortcuts such as the dataset
from where the image comes from than more complex
features such as lung opacities. While these shortcuts
lead to excellent performance in datasets similar to
the train dataset, catastrophic failure occurs once the
model is tested on a different dataset. Among others,
the markers for laterality, patient positioning and hos-
pital system were identified as features strongly in-
fluencing the decision of algorithms (DeGrave et al.,
2020).
The goal of this study was thus to develop and
validate an automatic method to detect markers and
written labels in CXR images. Such a method could
then be used for automatic obscuration of markers in
large datasets, promoting the learning of generic and
meaningful features and thus improving performance
and robustness.
2 METHODS
2.1 Datasets
Four different datasets were used in this study,
obtained from different sources. The first dataset,
hereinafter referred to as the Mixed dataset (1,395
CXRs) is composed of a combination of multiple pub-
lic CXR datasets, namely from the CheXpert (Irvin
et al., 2019) (7 CXRs), ChestXRay-8 (Wang et al.,
2017) (226 CXRs), Radiological Society of North
America Pneumonia Detection Challenge (RSNA-
PDC) (Kaggle, 2018) (639 CXRs) and COVID
DATA SAVE LIVES
1
(199 CXRs) datasets as
well as from COVID-19 CXR public repositories,
namely COVID-19 IDC (Cohen et al., 2020) (265
CXRs), COVIDx (Wang and Wong, 2020) (4 CXRs),
Twitter
2
(9 CXRs) and the Sociedad Espa
˜
nola de
Radiologia M
´
edica (SERAM) website
3
(46 CXRs).
The second and third datasets, hereinafter referred
to as the BIMCV and COVIGR datasets (289 and
300 CXRs respectively) are each from a single
hospital system public dataset, namely the BIMCV-
COVID19-PADCHEST (Bustos et al., 2020) (248
1
https://www.hmhospitales.com/coronavirus/
covid-data-save-lives
2
https://twitter.com/ChestImaging
3
https://seram.es/images/site/TUTORIAL CSI RX
TORAX COVID-19 vs 4.0.pdf
CXRs) and BIMCV-COVID-19+ (Vay
´
a et al., 2020)
(41 CXRs) datasets and the COVIDGR (Tabik et al.,
2020) dataset. The fourth dataset is a private col-
lection of 597 CXRs collected retrospectively at the
Centro Hospitalar de Vila Nova de Gaia e Espinho
(CHVNGE) in Vila Nova de Gaia, Portugal between
the 21st of March and the 22nd of July of 2020. All
data was acquired under approval of the CHVNGE
Ethical Committee and was anonymized prior to any
analysis to remove personal information.
All CXRs were selected randomly from both nor-
mal and pathological cases after exclusion of views
other than postero-anterior and antero-posterior.
2.2 CXR Annotation
In order to set a ground truth for training and evalu-
ation of the algorithms, manual annotation of all la-
bels was performed using an in-house software. The
software presented CXRs from a randomly selected
subset and allowed for window center/width adjust-
ment, zooming and panning. The software allowed
for rectangles of any size to be drawn on the image,
covering the labels, and saved the corresponding co-
ordinates. Figure 1 shows examples of manually an-
notated bounding boxes.
2.3 Automatic Label Detection
The automatic label detection model is based on
YOLOv3 (You Only Look Once, Version 3) (Red-
mon and Farhadi, 2018). The network is composed
of a feature extraction backbone, DarkNet-53 (Red-
mon and Farhadi, 2018), which is used to obtain
a M ×M ×N feature map F, where M is the spa-
tial grid used and N is the number of feature maps.
This feature map F is then convolved to obtain an
M×M×B×6 output tensor where B is a predefined
number of objects to predict per grid point and which
contains the predicted objects’ confidence score, class
probability and bounding box position and dimen-
sions. One particular characteristic of YOLOv3 is that
the bounding box dimensions are not explicitly pre-
dicted by the network but are defined in relation to
pre-defined bounding box templates, commonly re-
ferred to as anchors. The anchors are learned a pri-
ori before the training of YOLOv3 and correspond
to the cluster centers from a k-means that maximizes
the IoU of these anchors with the training set ground
bounding boxes. The network then learns to predict
the deviation (in length and width) from each of these
pre-defined anchors, thus defining each predicted ob-
ject.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
64
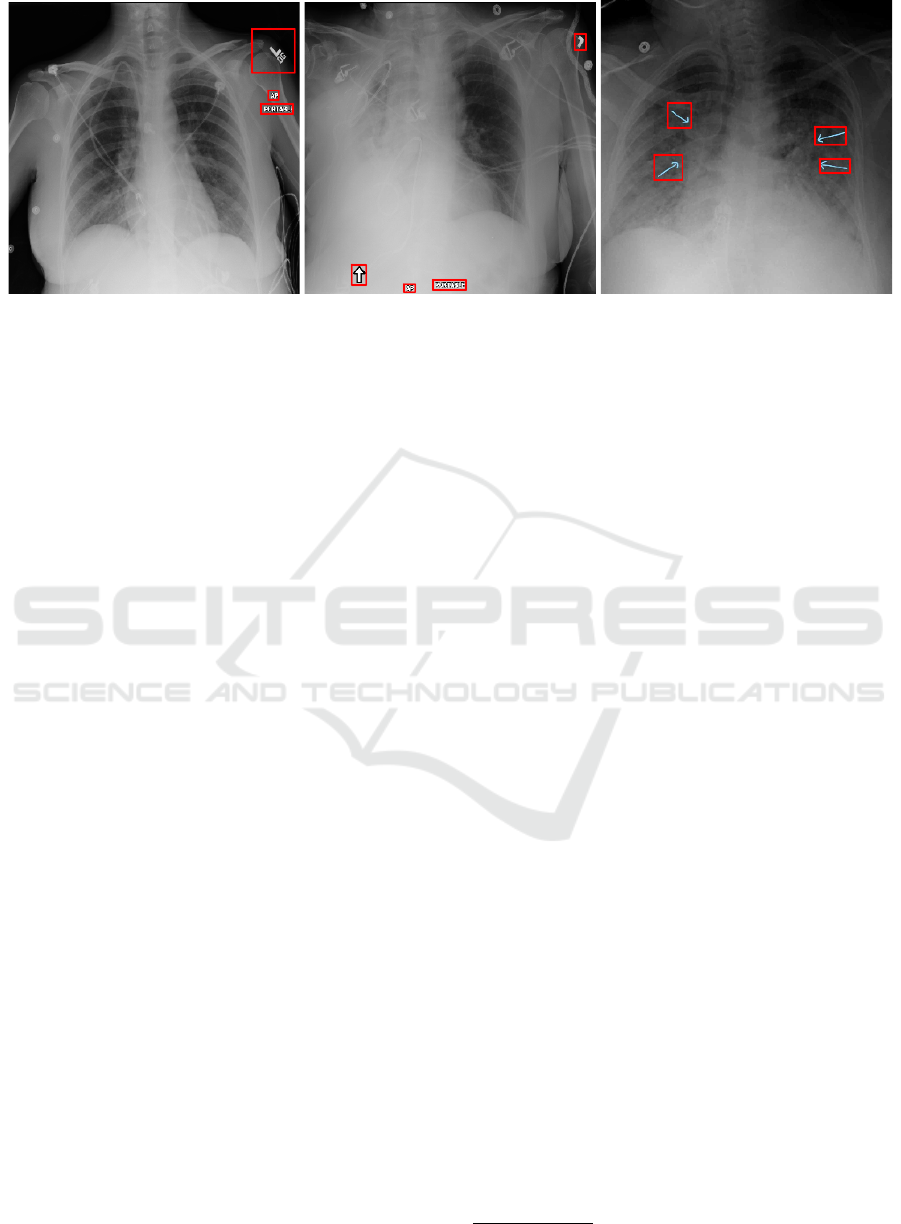
Figure 1: Example CXRs from the Mixed dataset with annotated bounding boxes covering equipment, laterality and patient
position markers among others.
In practice, this means that YOLOv3 divides the
image into a grid, defined according to M, and pre-
dicts objects for each image patch. For each object, a
confidence score and class probability are predicted,
as well as position and dimensions in relation to the
most similar anchor. During inference, predicted ob-
jects with low confidence score are then discarded to
obtain the final prediction.
3 EXPERIMENTS
3.1 CXR Annotation
Manual annotation was performed for all CXRs, re-
sulting in 1,202, 210, 656 and 1,298 annotated bound-
ing boxes for the Mixed, BIMCV, COVIDGR and
CHVNGE datasets respectively, which correspond to
an average of 1.51 bounding boxes per CXR.
3.2 Model Training
One quarter of the Mixed dataset was used for train-
ing/validation, ensuring that the same patient did not
appear in both training and testing. In total, 317 CXRs
(643 bounding boxes) were used for training and 39
CXRs (77 bounding boxes) were used for valida-
tion. The feature extraction backbone is the DarkNet-
53 (Redmon and Farhadi, 2018), and each cell has as-
sociated 9 anchor boxes. YOLOv3 pretrained weights
on MS-COCO (Lin et al., 2014) were used for initial-
ization. Training is performed with a batch size of 2,
Adam optimizer (Kingma and Ba, 2014) and learn-
ing rate of 10
−4
. The learning rate was lowered by
a factor of 10 whenever the validation loss did not
improve for 2 epochs and training was stopped if the
loss did not improve for 7 epochs. Data augmentation
was performed by applying random translations, flips
and scale changes. All experiments were conducted
on an Intel Core i7-5960X@3.00GHz, 32GB RAM,
2×GTX1080 desktop using Python 3.6, Tensorflow
2.0.0 and Keras 2.3.1.
3.3 Model Evaluation
Model evaluation was performed in terms of sensitiv-
ity and false positives (FP) per CXR. In a first ex-
periment, model predictions were compared to the
ground truth manual annotations for the Mixed test
set and the remaining datasets (BIMCV, COVIDGR
and CHVNGE). A prediction was considered a true
positive if it matches a ground truth bounding box.
Given that ground truth labels were obtained through
manual annotation and do not represent the minimum
bounding box for each label, a prediction was consid-
ered to be a match to a ground truth label if a coverage
of 40% was achieved.
In order to perform a more extensive validation
of the model, a subset of 100 CXRs was randomly
selected from all datasets and labels were then arti-
ficially placed in each CXR. Two additional experi-
ments were then conducted considering: 1) random
individual letters and 2) random English words
4
. For
each experiment, a total of 4,032 artificial labels were
placed per CXR, resulting in 403,200 artificially la-
beled CXRs per experiment. CXRs were divided into
8×8 quadrants and an equal number of labels was ran-
domly placed within each quadrant. Different label
font heights and intensity values were also consid-
ered, specifically font heights of 1/2, 1/4, 1/8, 1/16,
1/32, 1/64 and 1/128 in relation to CXR height and
relative intensity values of 0, 1/4, 1/2, 3/4, 1, 5/4, 3/2,
7/4 and 2. Label intensity values > 1 correspond to
artificial labels with intensity higher than the origi-
nal maximum image intensity. All parameters were
4
https://www.mit.edu/$\sim$ecprice/wordlist.10000
Automatic Label Detection in Chest Radiography Images
65
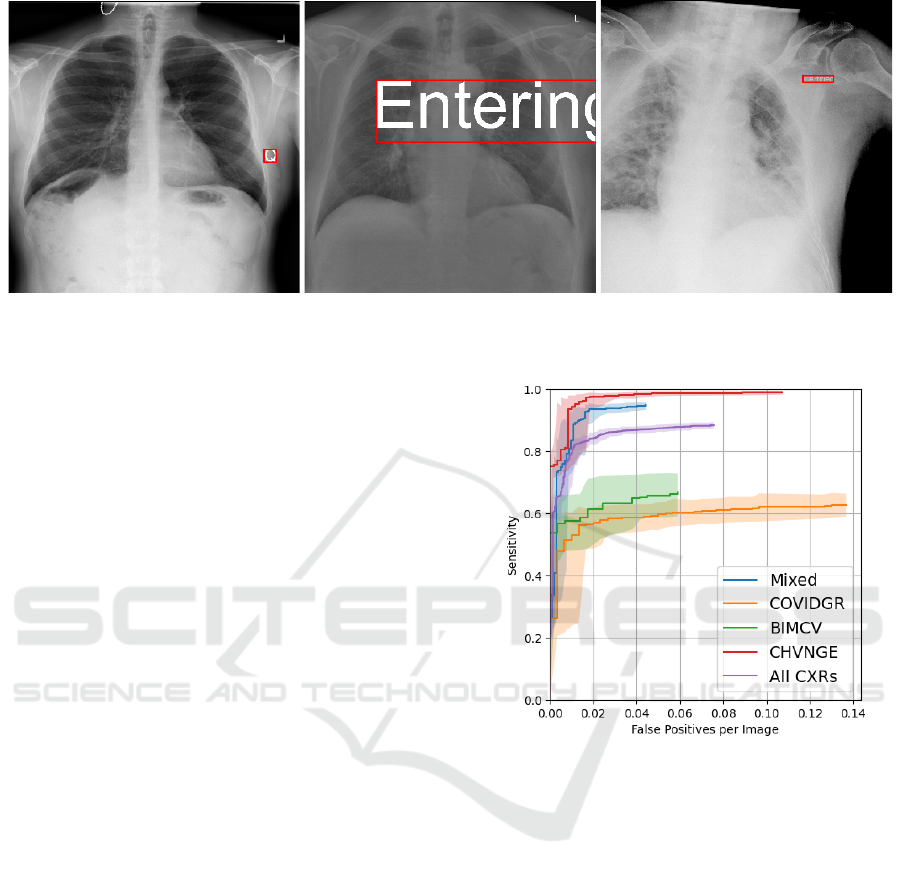
Figure 2: Example CXRs with artificially generated labels and corresponding minimum bounding boxes. (left) Label “Q” with
brightness 2 and font height 1/16; (middle) Label “Entering” with brightness 1.5 and font height 1/4; (right) Label “Certified”
with brightness 0.75 and font height 1/32.
selected empirically for an adequate representation
of label variability and sensitivity analysis. Figure 2
shows examples of artificially generated labels. Given
the goal of label obscuration, label coverage was also
used for evaluation for these experiments, defined as
the percentage of the area of the reference bounding
box covered by the predicted bounding box.
For all experiments, statistical error estimation
was performed by computing the 95% bias corrected
and accelerated (BC
a
) bootstrapping confidence in-
terval (CI) (Efron, 1987) calculated with 5.000 iter-
ations.
4 RESULTS
Figure 3 shows the free-response operating character-
istic (FROC) curve obtained for the ground truth la-
bels for all images and for each dataset, excluding the
CXRs used for training. It can be seen that a high
sensitivity is obtained for the Mixed and CHVNGE
datasets, while sensitivity for the COVIDGR and
BIMCV datasets saturates at approximately 0.6. All
datasets exhibit low FP rate per CXR. Figure 4 shows
examples of CXRs with ground truth and predicted
bounding boxes.
Figure 5 shows the sensitivity and coverage of the
artificial labels as a function of font height, relative
brightness and label position. Relative brightness was
computed as the difference between the artificial label
intensity and the average intensity of the CXR within
the minimum bounding box of the label.
Figure 3: FROC curve on all CXRs and each dataset.
Shaded region corresponds to the 95% BC
a
CI of each
FROC curve.
5 DISCUSSION
Figure 3 shows that a low number of FPs was
obtained, with high sensitivity, particularly for the
Mixed and CHVNGE datasets, in spite of the rela-
tively small training set used. As shown in Figure
4, the model was able to successfully distinguish be-
tween elements that belong to the original CXR and
those that were placed later. However, as illustrated
by the lower sensitivity obtained for COVIDGR and
BIMCV, the model struggled with faint and radio-
logical laterality markers (Fig. 4(b)) and particularly
small equipment markers (Fig. 4(c)) which were not
present in the training set.
Figure 5 further highlights the model capabilities,
showing that good sensitivity and coverage can be
obtained for both bright and dark labels, at differ-
BIOIMAGING 2022 - 9th International Conference on Bioimaging
66
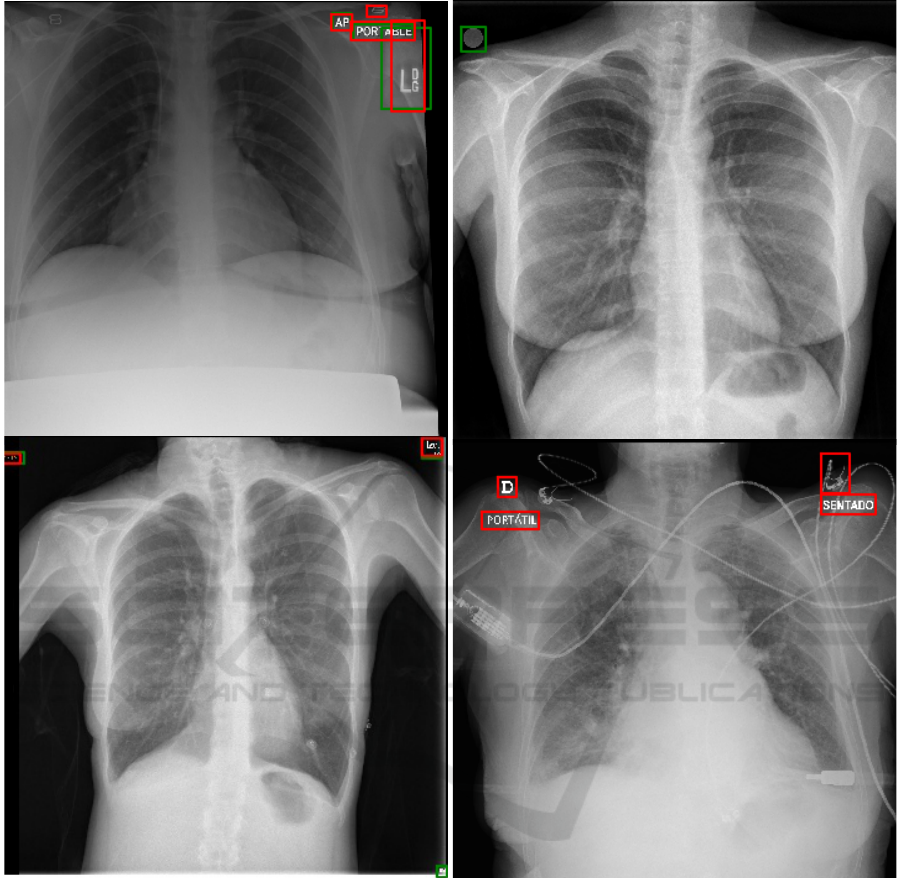
(a) (b)
(c) (d)
Figure 4: Example CXRs showing ground truth (green) and predicted (red) bounding boxes. (a) Mixed CXR; (b) BIMCV
CXR showing missed laterality marker; (c) COVIGR CXR showing missed equipment marker (right lower corner); (d)
CHVNGE CXR showing FP (medical device).
ent font sizes. As expected, subtle labels with rela-
tive brightness close to zero are more difficult to de-
tect, as well as extremely smalls labels at font sizes
under 1/32. Total label coverage is obtained for al-
most all font sizes, outside the more subtle relative
brightness ranges. Naturally, for extremely large font
sizes (over 1/8), coverage significantly drops as the
model struggles to identify words/letters as single la-
bels and instead predicts independent sections of the
label, thus failing to cover the significant portion of
CXR that falls within the label minimum bounding
box (Fig. 2 center). Surprisingly, slightly higher sen-
sitivity was observed for letters/words with negative
relative brightness for smaller objects (font height be-
low 1/8), which correspond to objects darker than the
background. Given that most of the ground truth an-
notations are of positive relative brightness, it can be
expected that the YOLOv3 has learnt to detect strong
edges, independent of the signal of the relative bright-
ness of the objects, which can be seen as proof of the
robustness of this method. Regarding the position of
the label, it can be seen that labels in the upper corners
can be detected more successfully, as most CXR la-
bels are placed in those regions, but reasonable perfor-
Automatic Label Detection in Chest Radiography Images
67
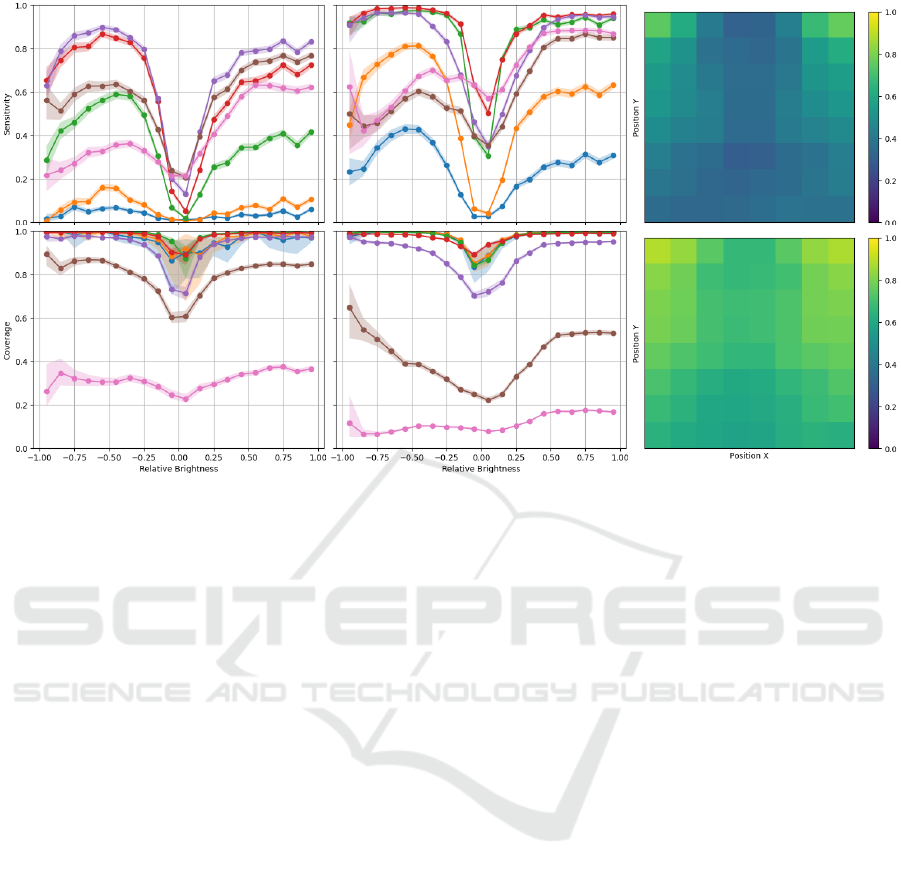
Figure 5: Sensitivity and coverage as a function of font height and relative brightness on the artificial letter (left) and word
(center) labels and sensitivity as a function of CXR position on the artificial letter (top right) and word (bottom right) labels.
Plot colors correspond to font height: • - 1/128; • - 1/64; • - 1/32; • - 1/16; • - 1/8; • - 1/4; • - 1/2. Shaded region
corresponds to the 95% BC
a
CI of each curve.
mance is nonetheless obtained in the remaining CXR
regions.
In spite of the promising results obtained, there are
limitations to this study which should be addressed.
A more thorough cross-dataset validation could be
performed and training with both manually annotated
and artificially generated labels could yield benefits
by improving performance for subtle and small labels
and for the lower portion of the CXR. Nevertheless,
in light of the low number of FPs per CXR and as
previously suggested, this framework could be used
to automatically obscurate markers in large datasets
with minimum oversight, potentially improving the
learning of generic and meaningful features. Alterna-
tively, it could also be used retrospectively in trained
models to infer whether shortcuts related to markers
have been learned by the model by computing the fre-
quency with which a model highlights image markers
as responsible for a given decision. Both these tech-
niques will be approached in future work.
6 CONCLUSION
In conclusion, an automatic CXR label detection
framework was proposed in this study based on a
YOLOv3 architecture. In spite of the relatively small
training set, it was shown that the model can be suc-
cessfully applied to datasets other than the training
data and good performance was shown for differ-
ent font sizes, relative brightness and position, being
thus an efficient method for label obscuration in large
datasets, potentially improving robustness and gener-
alization.
ACKNOWLEDGEMENTS
The authors would like to thank the Centro Hos-
pitalar de Vila Nova de Gaia/Espinho, Vila Nova
de Gaia (CHVNGE), Portugal for making the data
available which made this study possible. This
work was funded by the ERDF - European Regional
Development Fund, through the Programa Opera-
cional Regional do Norte (NORTE 2020) and by Na-
tional Funds through the FCT - Portuguese Foun-
dation for Science and Technology, I.P. within the
scope of the CMU Portugal Program (NORTE-01-
0247-FEDER-045905) and UIDB/50014/2020. Guil-
herme Aresta and Carlos Ferreira are funded by
the FCT grant contracts SFRH/BD/120435/2016 and
SFRH/BD/146437/2019 respectively
BIOIMAGING 2022 - 9th International Conference on Bioimaging
68

REFERENCES
Bustos, A., Pertusa, A., Salinas, J.-M., and de la Iglesia-
Vay
´
a, M. (2020). Padchest: A large chest x-ray image
dataset with multi-label annotated reports. Medical
Image Analysis, 66:101797.
Cohen, J. P., Morrison, P., and Dao, L. (2020). Covid-19 im-
age data collection. arXiv preprint arXiv:2003.11597.
DeGrave, A. J., Janizek, J. D., and Lee, S.-I. (2020). AI
for radiographic COVID-19 detection selects short-
cuts over signal. medRxiv.
Efron, B. (1987). Better bootstrap confidence inter-
vals. Journal of the American statistical Association,
82(397):171–185.
Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S.,
Chute, C., Marklund, H., Haghgoo, B., Ball, R., Sh-
panskaya, K., et al. (2019). CheXpert: A large chest
radiograph dataset with uncertainty labels and expert
comparison. In Proceedings of the AAAI Conference
on Artificial Intelligence, volume 33, pages 590–597.
Kaggle (2018). RSNA pneumonia detection chal-
lenge — kaggle. https://www.kaggle.com/c/
rsna-pneumonia-detection-challenge/. (Accessed on
10/07/2020).
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Lin, T.-Y., Maire, M., Belongie, S., Hays, J., Perona, P.,
Ramanan, D., Doll
´
ar, P., and Zitnick, C. L. (2014).
Microsoft coco: Common objects in context. In Euro-
pean conference on computer vision, pages 740–755.
Springer.
Qin, C., Yao, D., Shi, Y., and Song, Z. (2018). Computer-
aided detection in chest radiography based on artificial
intelligence: a survey. Biomedical engineering online,
17(1):113.
Redmon, J. and Farhadi, A. (2018). Yolov3: An incremental
improvement. arXiv preprint arXiv:1804.02767.
Tabik, S., G
´
omez-R
´
ıos, A., Mart
´
ın-Rodr
´
ıguez, J.,
Sevillano-Garc
´
ıa, I., Rey-Area, M., Charte, D.,
Guirado, E., Su
´
arez, J., Luengo, J., Valero-
Gonz
´
alez, M., et al. (2020). COVIDGR dataset and
COVID-SDNet methodology for predicting COVID-
19 based on chest x-ray images. arXiv preprint
arXiv:2006.01409.
Vay
´
a, M. d. l. I., Saborit, J. M., Montell, J. A., Pertusa,
A., Bustos, A., Cazorla, M., Galant, J., Barber, X.,
Orozco-Beltr
´
an, D., Garcia, F., et al. (2020). BIMCV
COVID-19+: a large annotated dataset of RX and
CT images from COVID-19 patients. arXiv preprint
arXiv:2006.01174.
Wang, L. and Wong, A. (2020). COVID-Net: A tailored
deep convolutional neural network design for detec-
tion of COVID-19 cases from chest x-ray images.
arXiv preprint arXiv:2003.09871.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and
Summers, R. M. (2017). Chestx-ray8: Hospital-
scale chest x-ray database and benchmarks on weakly-
supervised classification and localization of common
thorax diseases. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition,
pages 2097–2106.
Automatic Label Detection in Chest Radiography Images
69
