
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting
Multi-skin Cancer Diseases over Dermoscopic Images
Robert Brodin, Palawat Busaranuvong and Chun-Kit Ngan
Data Science Program, Worcester Polytechnic Institute, Worcester, MA, U.S.A.
Keywords: Convolutional Neural Network, Skin Cancer Detection, Automatic Architecture Design, Dermoscopic Image.
Abstract: We enhance and customize the automatically evolving genetic-based CNN (AE-CNN) framework to develop
an auto-designed CNN (AutoCNN) pipeline to dynamically generate an optimal CNN model to assist
physicians in detecting multi-skin cancer diseases (MSCD) over dermatoscopic images. Specifically, the
contributions of this work are three-fold: (1) integrate the pre-processing module into the existing AE-CNN
framework to sanitize and diversify dermatoscopic images; (2) enhance the evaluation algorithm of the
framework to improve the model selection process by using the k-fold cross-validation; and (3) conduct the
experimental study to present the accuracy results that the CNN model constructed by AutoCNN outperforms
the model by AE-CNN to detect and classify MSCD.
1 INTRODUCTION
Skin Cancer is one of the fastest-growing diseases in
the United States, and most commonly is the
abnormal growth of skin cells with the ability to
spread to neighboring cells or other parts of the body.
According to the American Academy of Dermatology
Association (AADA, 2021), there are approximately
9,500 people in the U.S. diagnosed with skin cancer
every day (i.e., an average of 3.3 million Americans
per year). Despite this, if skin cancer can be detected
and diagnosed early, the five-year survival rate for
patients is expected to be 98 percent (ACS, 2021). To
support physicians to detect and diagnose skin cancer
early in its development for patients, dermatoscopy
(DermNet, 2021) is a widely used technique, as
dermatoscopic images have an immense potential for
the detection of a suspicious mole at an early stage of
development. However, for physicians to detect and
classify a skin cancer early into a specific type (e.g.,
melanoma, melanocytic nevus, basal cell carcinoma,
actinic keratosis, and benign keratosis) on
dermatoscopic images is very challenging due to
various subjective and time-intensive interpretations.
To address the above issues, convolutional neural
networks (CNNs) have played an important role to
speed up the early detection of skin cancer through
dermatoscopic image classification. In a study
comparing the detection accuracy of CNNs with
trained dermatologists, CNNs were found to be
72.1% accurate, as opposed to 65.56%-66% with
trained dermatologists (Chan, S., 2020). Using this
state-of-the-art technology to support the early
detection of skin cancers, physicians have a more
effective and efficient way to treat their patients. The
more effective and efficient detection and diagnosis
of skin cancer improves the survival rate of patients.
Therefore, we are motivated to study and explore this
technology to address the detection of skin cancer.
Presently, the construction of CNN architectures
to conduct the early detection of skin cancers can be
broadly divided into two categories: single and
ensemble. Built on domain knowledge and the
available datasets, single hand-crafted CNNs
(Albelwi, S, 2016; Nasr-Esfahani, E., 2016; Le, T.T.,
2017; Stefan Jianu, S.R., 2019; Ashraf Ottom, M.
2019; Rundo, F., 2019; Fu’adah, Y.N., 2020) are the
manually-designed static architectures that require
researchers to possess significant medical domain
knowledge and deep CNN design experience to
develop and implement the network. With
considerable domain expertise, the best CNN
architecture constructed by this approach delivers a
certain promising detection performance on the
dataset originated from homogeneous data sources.
However, due to its insufficient and imbalanced data
to train the network, the architecture may not behave
well for unseen types of data images from
heterogeneous data sources, as in many cases, the
Brodin, R., Busaranuvong, P. and Ngan, C.
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting Multi-skin Cancer Diseases over Dermoscopic Images.
DOI: 10.5220/0010893400003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
607-615
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
607

network is not able to learn enough representative
instances for each class label (i.e., skin cancer types)
to extract the distinctive image features for the
classification. In addition, those heterogeneous
datasets may exhibit different data variations and
characteristics that the hand-crafted CNNs may not be
able to provide promising detection performance.
To address these issues, ensemble CNN
architectures have been developed. Specifically, they
are hybrid CNN architectures (Aldwgeri, A., 2019;
Mahbod, A., 2019; Al Mamun, Md., 2021) that
combine different single CNN models, which have
been pre-trained on a large number of images and
adapted to their diverse variations to extract more
unique features from the domain-specific images.
Models that conduct ensemble learning deliver a better
classification performance than that of single CNN
models only. Currently, there are several well-known
pre-trained CNN models, such as VGGNet (Simonyan,
K., 2015), GoogleNet (Szegedy, C., 2015), and ResNet
(He, K., 2016), which are pre-trained on ImageNet
(ImageNet, 2021) and CIFAR (Krizhevsky, A., 2009).
However, to identify the best possible combination of
those pre-trained CNN models is challenging due to a
large number of different possible model combinations
with the high computational learning cost. In addition,
even though learning the best model combination
among all the possible pre-trained CNNs is a dynamic
process, those pre-trained CNN models are still in the
static architectures that may lack the adaptability for
diverse image variations (e.g., skin cancer images).
Those pre-trained CNN models may perform
significantly worse with heterogeneous data sources
not encountered before.
To bridge the above gaps, (Sun, Y., 2020) have
developed an automatically evolving genetic-based
CNN (AE-CNN) framework to dynamically design
and construct an optimal CNN architecture on any
available image dataset without requiring any manual
intervention. The experimental results show that the
CNN architecture generated from the framework
outperforms the above state-of-the-art CNNs’ peer
competitors in terms of the classification accuracy
performance. However, the AE-CNN framework that
generates an optimal architecture is not fully designed
for skin cancer detection and classification and does
not consider two crucial components: (1) pre-
processing the raw images (e.g., lesion segmentation,
image augmentation, etc.,) and (2) selecting the best
CNN model based upon the entire training dataset
instead of a separated validation dataset only. To
mitigate the above shortcomings, we enhance and
customize AE-CNN to develop and implement an
auto-designed CNN (AutoCNN) framework that
enables domain users to dynamically generate an
optimal CNN architecture on their available datasets
to assist physicians in early detecting multi-skin
cancer diseases (MSCD) over dermatoscopic images.
Specifically, the contributions of this work are three-
fold: (1) integrate the pre-processing module into AE-
CNN to sanitize and diversify dermatoscopic images,
(2) enhance the evaluation algorithm of AE-CNN to
improve the model selection process by using the k-
fold cross-validation (Sanjay, M., 2018) on the entire
training dataset, and (3) conduct an experimental
study, using the 25,331 dermatoscopic images
provided by the 2019 International Skin Imaging
Collaboration (ISIC, 2019), to present the
classification accuracy. From the results, we can
conclude that the CNN model constructed by
AutoCNN outperforms the model constructed by AE-
CNN to detect and classify MSCD. The source code
will be available to the public after the acceptance.
The remainder of the paper is organized as
follows. First, we briefly describe the AE-CNN
framework in Section 2. In Section 3, we illustrate our
enhanced AutoCNN framework and its workflow.
We also demonstrate and explain our pipelines of
both skin cancer image segmentation and its
augmentation in Section 4 and 5, respectively. In
Section 6, we discuss and summarize the
experimental study and results. In Section 7, we
conclude and briefly outline our future work.
2 AE-CNN FRAMEWORK
Fig. 1 is the AE-CNN framework. First, the size of
the population N, i.e., the total number of individual
CNN architectures in each generation, is predefined.
Note that each CNN individual in each generation is
trained on 80% of the ISIC-2019 image dataset and
validated on 10% of the dataset in the fitness
evaluation module in our experimental study. Each
CNN individual of a generation in the population N
takes part in the evolutionary process of the genetic
algorithm with the maximal generation number of T.
During the population evolutionary process, a new
CNN offspring is generated from its selected parents
with the crossover and mutation operations, while the
parents are selected by the binary tournament
selection. After the fitness of the generated offspring
has been evaluated, a new population is selected with
the environmental selection operation from the
current operation that contains the current individuals
and the generated offspring, and the parents survive
into the next evolutionary process, the next
generation. Towards the end, the framework
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
608
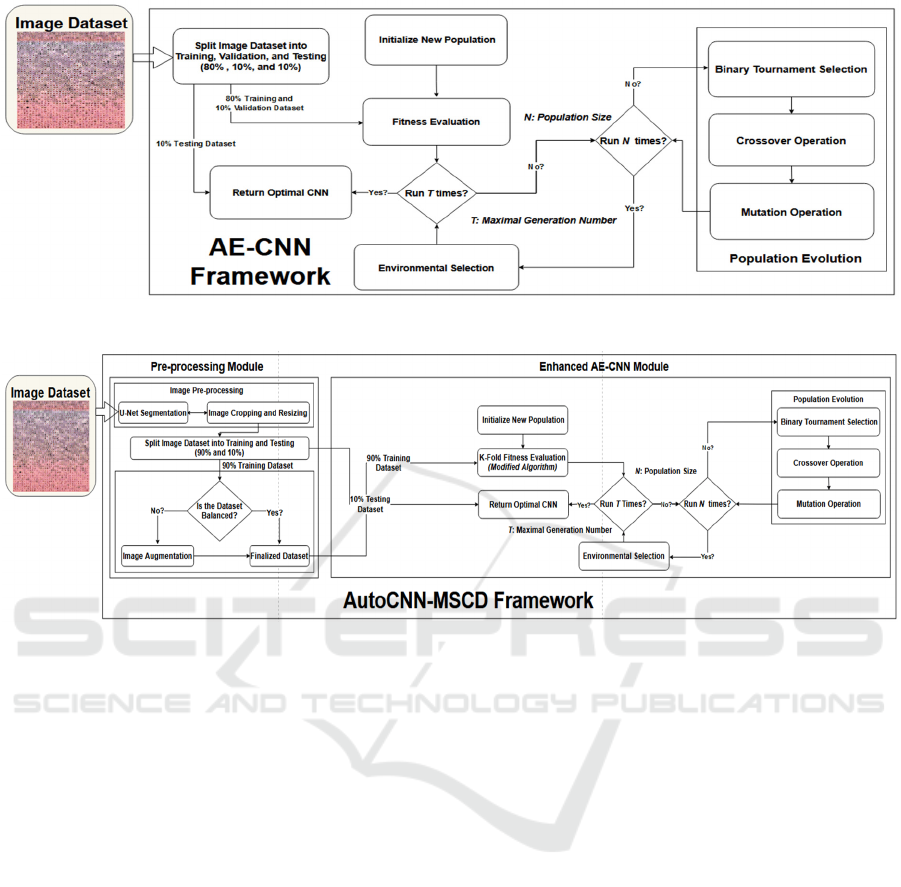
Figure 1: Automatically Evolving CNN (AE-CNN) Framework.
Figure 2: Autodesigned CNN Framework for Multi-Skin Cancer Diseases (AutoCNN-MSCD) Detection.
generates T * N CNN models, from which the best
CNN architecture is selected among all the possible
CNN candidates in terms of their classification
accuracy performance. The best CNN model
generated by AE-CNN is then compared with the best
model constructed by AutoCNN. Both CNN models
are tested on the same 10% of the image dataset for
performance evaluation.
3 AutoCNN-MSCD FRAMEWORK
Fig 2. is the AutoCNN framework, which is the
enhanced version of AE-CNN to detect MSCD. The
framework is composed of two main modules: Pre-
processing and Enhanced AE-CNN. In the pre-
processing module, there are three sub-modules
including Image Resizing (IR), U-Net Segmentation
(Ronneberger, O., 2015), and Image Cropping (IC).
First, the framework loads the raw images of each class
label into the IR sub-module to resize each image. The
resized images are then passed to the U-Net sub-
module that performs the semantic segmentation
process to locate the position of skin lesions on the
images. After that, the skin lesions on the images are
cropped and resized again. The cropped and resized
images per class label are randomly split into 90% for
training and 10% for testing. Each skin cancer class in
the 90% training dataset is iterated through and the
number of images is counted. If the quantities are not
balanced among all the classes, then image
augmentation is conducted to generate enough images
for each of the classes. The goal is to ensure that each
CNN model per generation gets trained equally on each
class of skin cancer. The 10% testing dataset is not
augmented to prevent data leakage and overfitting
problems. After the images are segmented and
augmented, they are loaded into the enhanced AE-
CNN module. The main difference between the
original AE-CNN framework and the enhanced AE-
CNN module is that the original AE-CNN framework
evaluates each CNN model only on a specific portion
of the original dataset. The accuracy calculated is not
fully complete, whereas the enhanced AE-CNN
module assesses the performance of each CNN
individual by using 10-fold cross-validation. 10-fold
cross-validation gives a more accurate performance of
the best CNN model among all possible candidates
(Gholamiangonabadi, D., 2020; Shaban, M., 2020).
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting Multi-skin Cancer Diseases over Dermoscopic Images
609
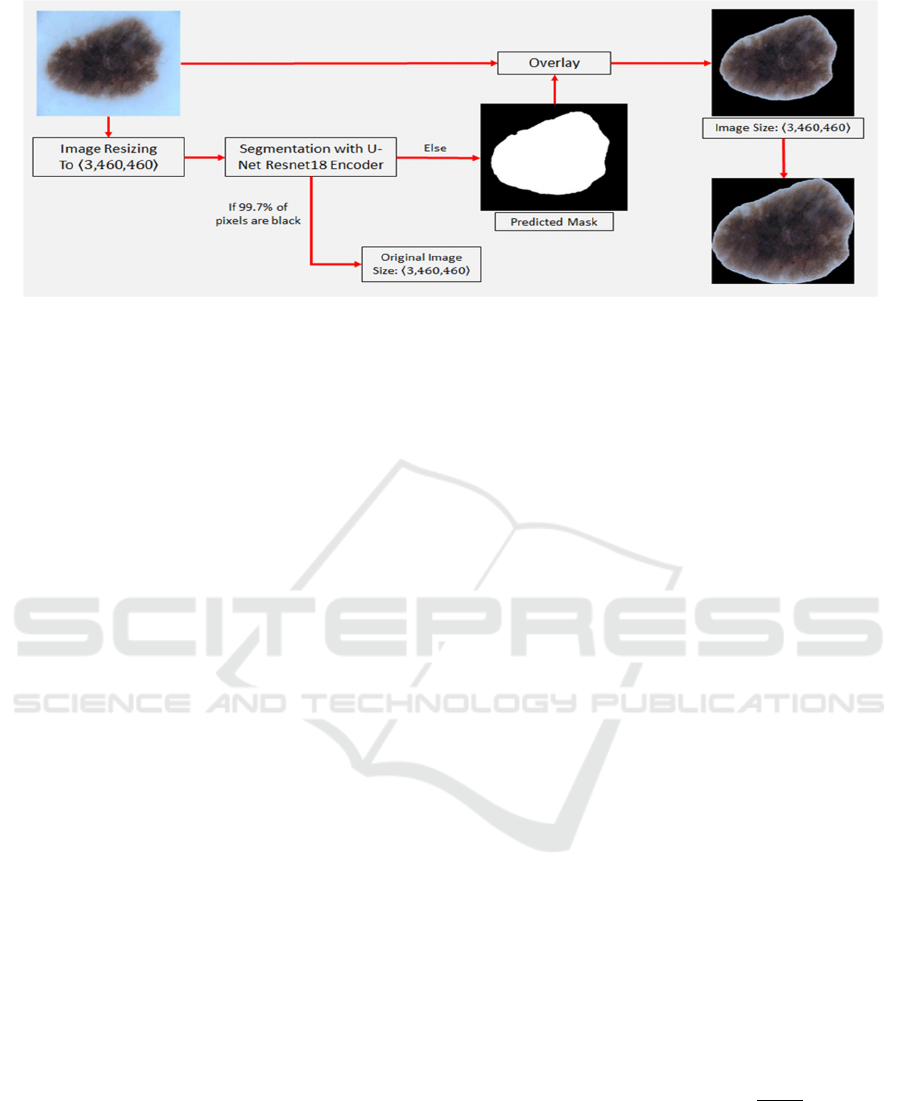
Figure 3: Image Pre-processing Module.
4 IMAGE PRE-PROCESSING
MODULE
In this section, we describe and explain the pre-
processing module in more detail. Some images may
have a lesion at the center/border or some may have a
tiny tumor on a specific spot of the photo, which is
very difficult for a CNN model to detect. Therefore,
we have developed and implemented the image pre-
processing module to locate a lesion on each image
and then crop the area not including the lesion from
the image. Each CNN model can learn each type of
lesions more precisely rather than learning its
surrounding background with noise.
Our image pre-processing pipeline is shown in Fig
3. First, each skin cancer image is resized to 460 x
460 pixels in the RGB channels. After that, the
resized images are passed to the U-Net architecture
with the ResNet18 encoder to obtain the predicted
mask of each image lesion. The predicted mask is
then overlayed on top of the resized images. This
overlayed image is cropped and resized again so that
the lesion is in the middle of the image. However,
since some predicted masks are almost completely
dark, i.e., more than 99.7% of the mask is entirely
occupied by the black pixels, the original image is
then used for the downstream process without
performing the segmentation. In this case, a mask
cannot provide any information about the lesion on
the image other than a completely dark photo.
4.1 U-Net Architecture with ResNet18
Encoder
In the image pre-processing module, we use the U-
Net architecture to perform the skin lesion semantic
segmentation task, because the U-Net architecture,
with several plain convolutional layers, is originally
designed and developed for biomedical image
segmentation. Despite this, it is not completely used
for skin cancer segmentations. Due to the flexibility
of the FastAI library (FastAI, 2021), we have
replaced the down-sampling section of the original U-
Net encoder with another state-of-the-art CNN
architecture, ResNet. The ResNet network is selected
because of its advantage of being able to skip
connections to prevent overfitting, to avoid a
vanishing gradient, and to extract more important
features than the plain convolutional layers. The
enhanced U-Net decoder is similar to the basic U-Net
decoder, which also has cross-connections. The main
difference is that instead of up-sampling by using
transpose convolutions, FastAI applies a newly
developed method, i.e., pixel shuffle or sub-pixel
convolution with ICNRN initialization, which is used
in the image’s super-resolution (Shi, W., 2016).
To train and evaluate the enhanced U-Net with a
ResNet encoder, we use the ISIC-2018 dataset (ISIC,
2018), which contains 2,594 training images with
their corresponding ground-truth masks, because the
ISIC-2019 dataset does not contain ground-truth
masks. Specifically, we use 90% of the images for
training and 10% for testing. During the training
process, we select two different ResNet backbones,
i.e., ResNet18 and ResNet34. Note that our
framework can flexibly apply to any backbones based
upon the image sources and variety. The evaluation
metric is the Dice Coefficient (DC) score computed
by using this equation: DC Score =
|∩|
||||
, where G is
a set of ground truth images, P is a set of segmented
images, |G| and |P| are the number of images in each
set, and |G∩P| is the number of intercepted images.
The DC score measures the similarity of the ground
truth images and the predicted images by the U-Net
with the two ResNet encoders.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
610

Figure 4: Skin Lesion Semantic Segmentation and Ground Truth on the ISIC-2018 Testing Dataset.
The U-Net training procedure is as follows: (1)
apply the pretrained weights of the ResNet encoder
and freeze the encoder weights, (2) train the weights
of the U-Net decoder with the Adam optimizer with
the batch size of 8 and the learning rate of 0.001 for
10 epochs, and (3) unfreeze the weights of the
encoder section and continue training the whole
network with the same optimizer but using a smaller
learning rate for 10 epochs. To reduce the overfitting
of training the whole network, we apply the early
stopping by truncating the training process when the
validation DC score is not improved for three epochs
and take the optimal weights from the epoch with the
highest validation score.
Fig. 4 shows some examples of the U-Net
semantic segmentation on the ISIC-2018 testing
dataset, in which we cannot find a big difference
between the ResNet34 and ResNet18 backbones, in
terms of their segmentation capability. Table 1 also
shows the segmentation results on the ISIC-2018
testing images, where we observe that the DC score
of the ResNet34 encoder is slightly better than that of
the ResNet18 encoder. Note that the size of a lesion
is relatively big in each image and the position of the
lesion is mostly at the center of the images in the
ISIC-2018 dataset. On the contrary, the ISIC-2019
skin cancer images contain eight different types of
lesions (including melanoma, melanocytic nevus,
basal cell carcinoma, actinic keratosis, benign
keratosis, dermatofibroma, vascular lesion, and
squamous cell carcinoma) and their locations are not
always at the center/border of the images. Because of
this, we evaluate the U-Net architecture with both
ResNet encoders to see which one is more capable of
handling this problem. We found that the U-Net
architecture with ResNet34 and ResNet18 encoders
both can detect the edges of the majority of lesions on
the ISIC-2019 images. However, for the small lesions
on the images, the U-Net with the ResNet18 encoder
can still detect the lesions, while the U-Net with the
ResNet34 encoder fails to capture any lesion on the
images. Some examples are shown in Fig. 5. Due to
the performance of the ResNet18 encoder, which can
spot small lesions on the image, and its DC score
which is slightly lower than that of the ResNet34
encoder, we decide to integrate it into our U-Net
architecture.
Table 1: Dice Coefficient Score on Testing Images.
Encoder Name Dice Coefficient Score
U-Net with RestNet18 0.864
U-Net with RestNet34 0.878
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting Multi-skin Cancer Diseases over Dermoscopic Images
611
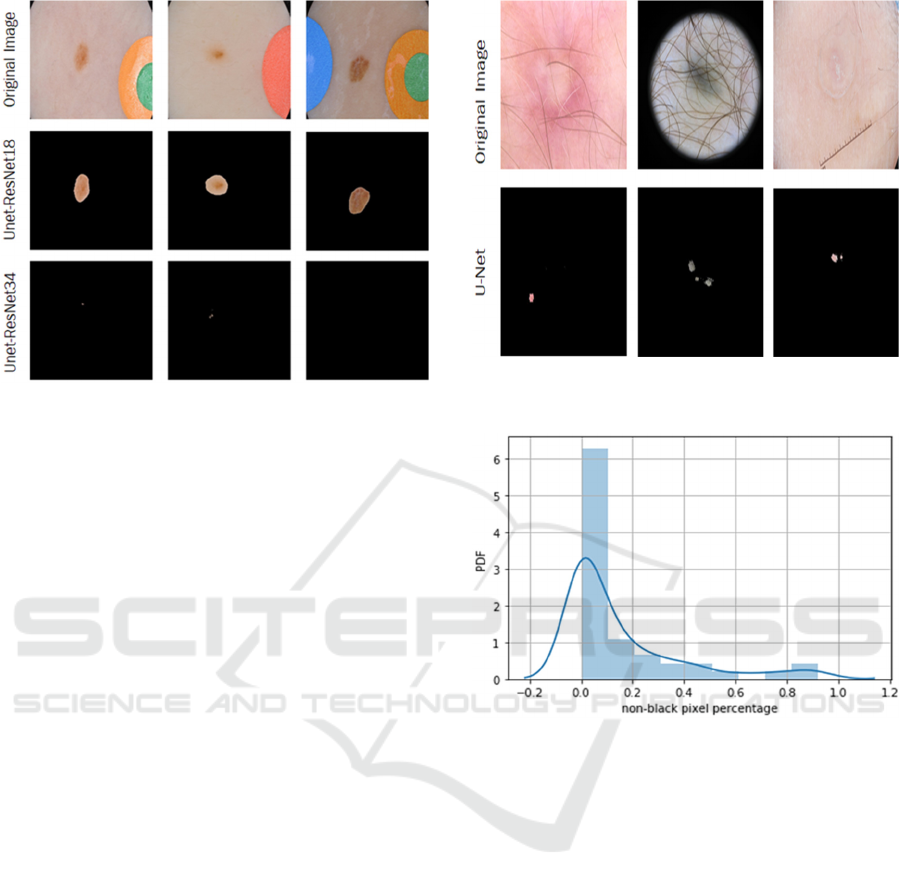
Figure 5: Examples that the U-Net with ResNet34 Encoder
Fails to Detect a Lesion on the ISIC-2019 images.
4.2 Threshold Determination to Use
Original Images for CNN Training
Although the ResNet18 encoder can perform better
than the ResNet34 encoder for spotting small lesions
on images, it still cannot detect irregular and
unobvious lesions on images shown in Fig. 6. Note
that among 25,531 ISIC-2019 images, there are 45
images that the ResNet18 encoder cannot detect that
results in generating almost completely dark
segmented images. For those images, we do not
perform the segmentation because there is no
advantage of using those completely dark images to
train our CNN models. Instead, we just resize those
original images as the same size as the other
segmented images.
To make the pipeline automatically identify this
type of image, shown in Fig. 6, which cannot be
detected by the ResNet18 encoder, we need to set a
proper threshold as a cut-off point. First, we analyze
the characteristics of those 45 images, as a base, by
computing the percentage of the non-black pixels in
those images. The percentage distribution is shown in
Fig. 7 that the mean value is 0.153%, the median
value is 0.030%, and the standard deviation (SD) is
0.236%. As the distribution is more right-skewed,
most of the images have very small non-black pixels.
To detect those image outliers, we decide to select a
threshold using the average of Median + SD (0.27%)
and Mean + SD (0.39%), i.e., 0.3%, in the dataset. It
means that if a segmented image contains the
percentages of the black pixels at least 99.7% or
more, we use its original image for the down-
streaming processes for the CNN training.
Figure 6: Examples that the U-Net with the ResNet18
Encoder Fails to Detect a Lesion on the ISIC-2019 images.
Figure 7: Distribution Plot of Non-black Pixel Percentage.
5 IMAGE AUGMENTATION
MODULE
The collected images that AE-CNN and AutoCNN
are evaluated on is the ISIC-2019 skin cancer dataset,
which includes eight different skin cancer diseases.
The types of skin cancer are melanoma, melanocytic
nevus, basal cell carcinoma, actinic keratosis, benign
keratosis, dermatofibroma, vascular lesion, and
squamous cell carcinoma. The number of images in
each disease class varies from 250 to 10,000 images
that the augmentation is needed to make the image
balance among the classes to improve the
performance of CNN models. Our image
augmentation pipeline first finds the class with the
largest number of images and then augments the
images in each other class until it equals the number
of images of the largest class. Our pipeline includes
four different image augmentation methods, shearing,
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
612
flipping, random contrast, and random noise,
conducted in sequence, shown in Table 2. Note that
the augmentation pipeline is programmed in Python
using a combination of two libraries, Augmentor
(Bloice, M.D., 2020) and Albumentations
(Albumentations Team, 2021). Both libraries have
different types of image augmentations and are
applied in different ways. The Augmentor library
used in the augmentation pipeline is to perform the
shearing, flipping, and random contrast operations,
while the Albumentations library is used to apply the
random noise augmentation. The number of
augmentations needed is calculated for each class and
the functions of these specific augmentations are
called to produce the new images.
Table 2: Image Augmentation Techniques.
Method Probability Parameters
Shearing 1.0
Max_shear_left = 25
Max_shear_right = 25
Flipping 0.8 NA
Random
Contrast
0.8
Min_factor = 0.9
Max_factor = 1.0
Random Noise 1.0
Mode = 's&p'
Amount = 0.02
In our setting, for each randomly selected image,
there are at least two or up to four augmentation
methods being used that depends on the probability
of that method being set. First, an image from each
other class is randomly selected and then the shearing
augmentation is conducted on the image due to the
100% probability of occurring. After that, the image
may or may not be conducted on the flipping
augmentation because of its 80% probability
happening. This process continues for the other two
augmentations in order. Each augmentation function
also has its own specific parameters. Flipping has no
parameters. Shearing has a max rotation to the left
and a max rotation to the right, e.g., the max angles
are not more than 25 degrees in each rotation.
Random contrast has the minimum and maximum
factor parameters (e.g., 0.9 and 1.0 respectively),
which corresponds to the lower and upper bound of
how much contrast is contained in an image. Random
noise has two parameters: "mode" decides what type
of noises is applied (e.g., salt and pepper (s&p)) and
"amount" determines the percentage of image pixels
containing noise (e.g., 0.02). After the process is
completed for each other class, the augmented images
are moved into their own class folders.
6 EXPERIMENTAL RESULTS
AND DISCUSSION
Based upon the classification accuracy performance
among all the eight skin cancer diseases in the 10%
validation dataset performed by each CNN individual
among all the 15 generations in the AE-CNN
framework, we find that the 3
rd
individual in the 14
th
generation, shown in the Table 3 configuration, has
the highest validation accuracy with 67.12%. This
CNN model is composed of six main units in order,
where DBU is a DenseNetUnit, RBU is a
ResNetUnit, and APL is an Average Pooling Layer.
In each unit, there is its own specific configuration.
For example, the 1
st
DBU has three feature filters (F)
with the input dimension (I) 32 x 32 pixels. After an
image is passed through the 1
st
DBU, there are three
corresponding feature maps generated as the outputs.
The size of each feature map (O) is 16 x 16 pixels.
Table 3: Best CNN Model Configuration from AE-CNN.
1
st
2
nd
3
rd
4
th
5
th
6
th
DBU DBU DBU RBU RBU APL
F: 3
I: 32x32
O: 16x16
F: 6
I: 16x16
O: 12x12
F: 12
I: 12x12
O: 32x32
F: 3
I: 32x32
O: 32x32
F: 1
I: 32x32
O: 1x8
Average
Pooling
Likewise, using the 10-fold cross-validation
accuracy of each CNN model in AutoCNN, we notice
that the 2
nd
individual in the 15
th
generation, shown in
the Table 4 configuration, has the highest accuracy
(70.82%) on the 90% training dataset. This CNN
model is constructed by seven main units in order.
Table 4: Best CNN Model Configuration from AutoCNN.
1
st
2
nd
3
rd
4
th
5
th
6
th
7
th
RBU DBU RBU RBU DBU RBU APL
F: 4
I: 32x32
O: 39x39
F: 8
I: 39x39
O: 24x24
F: 16
I: 24x24
O: 16x16
F: 3
I: 16x16
O: 32x32
F: 4
I: 32x32
O: 32x32
F: 1
I: 32x32
O: 1x8
Average
Pooling
Both individuals above are selected and evaluated
on the testing dataset. Note that the main reason why
only five individuals in 15 generations in each
framework are executed is that we lack the
computational hardware to execute both AE-CNN
and AutoCNN frameworks. We used the WPI Turing
Research Cluster (WPI, 2018), which limits each job
to a certain number of resources over a certain amount
of time. To run both frameworks to generate and train
each CNN individual, our jobs are limited to two
NVIDIA P100 graphics cards, 2 Intel CPUs, and
approximately 50 gigabytes of memory. Due to these
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting Multi-skin Cancer Diseases over Dermoscopic Images
613

limitations, we are only able to generate and train five
CNN models among all the 15 generations. These
computation limitations cause both frameworks to
take a significant amount of time to run to
completion, approximately one week each. If we had
accessed to more resources, we would have been able
to run more individuals and more generations to
obtain a higher-performing CNN model. Despite
these, once the best CNN model is obtained, the skin
cancer detection is very fast. The testing result shown
in Table 5 suggests that the best CNN model of
AutoCNN outperforms the best of AE-CNN by about
5.23% for eight different classes of skin cancer
diseases. AutoCNN has higher accuracy, because of
the image pre-processing module and the k-fold
cross-validation approach. Our results have shown a
promising improvement in AutoCNN.
Table 5: Testing Accuracy of AE-CNN and AutoCNN Best
Generation
Model Generation Individual Accuracy
AE-CNN 14 3 0.6642
AutoCNN 15 2 0.7165
7 CONCLUSIONS AND FUTURE
WORK
To the best of our knowledge, this is the first paper
written to enhance and customize genetic-based AE-
CNN to develop and implement an AutoCNN
framework that enables domain users to dynamically
generate an optimal CNN architecture on their
available datasets to assist physicians in early
detecting MSCD over dermatoscopic images.
Specifically, the contributions of this work are three-
fold: (1) integrate the pre-processing module into AE-
CNN to sanitize and diversify dermatoscopic images,
(2) enhance the evaluation algorithm of AE-CNN to
improve the model selection process by using the k-
fold cross-validation on the entire training dataset,
and (3) conduct an experimental study, using the
25,331 dermatoscopic images provided by ISIC-
2019, to present the classification accuracy. From the
results, we can conclude that the CNN model
constructed by AutoCNN outperforms the model
constructed by AE-CNN to detect and classify
MSCD. However, there are still several important
research challenges that we would like to investigate
into the performance of AutoCNN concerning
MSCD. Specifically, how the metadata of each
image, including the patient's age, gender, and lesion
location, can impact the classification accuracy of
CNN models. We are also interested in how Deep-Q
learning algorithms can improve the speed of
generating CNN architectures, as well as how the
framework can be enhanced to provide a user-friendly
interface for domain users to easily operate it.
ACKNOWLEDGEMENTS
This work has been partially supported by the 2020
Early Research Experience in E-Term (EREE)
Program at Worcester Polytechnic Institute.
REFERENCES
Albelwi, S. and Mahmood, A. (2016). Automated Optimal
Architecture of Deep Convolutional Neural Networks
for Image Recognition. The 15
th
IEEE International
Conference on Machine Learning and Applications.
doi: 10.1109/ICMLA.2016.0018.
Aldwgeri, A. and Abubacker, N.F. (2019). Ensemble of
Deep Convolutional Neural Network for Skin Lesion
Classification in Dermoscopy Images. In: Badioze
Zaman H. et al. (eds) Advances in Visual Informatics.
IVIC 2019. Lecture Notes in Computer Science, vol
11870. Springer, Cham. https://doi.org/10.1007/978-3-
030-34032-2_20.
Al Mamun, Md. and Uddin, M.S. (2021). Hybrid
Methodologies for Segmentation and Classification of
Skin Diseases: A Study. Journal of Computer and
Communications, 9, 67-84. doi: 10.4236/jcc.2021.94
005.
Albumentations Team. (2021). Fast image augmentation
library and an easy-to-use wrapper around other
libraries. https://github.com/albumentations-team/
albumentations.
American Academy of Dermatology Association (AADA).
(2021). Skin Caner. https://www.aad.org/media/stats-
skin-cancer.
American Cancer Society (ACS). (2021). Survival Rates
for Melanoma Skin Cancer. https://www.cancer.org/
cancer/melanoma-skin-cancer/detection-diagnosis-
staging/survival-rates-for-melanoma-skin-cancer-by-
stage.html.
Ashraf Ottom, M. (2019). Convolutional Neural Network
for Diagnosing Skin Cancer. The International Journal
of Advanced Computer Science and Applications, Vol.
10, No. 7.
Bloice, M. D. (2020). Image augmentation library in Python
for machine learning. https://github.com/mdbloice/
Augmentor.
Chan, S., Reddy, V., Myers, B., Thibodeaux, Q.,
Brownstone, N., & Liao, W. (2020). Machine Learning
in Dermatology: Current Applications, Opportunities,
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
614

and Limitations. Dermatology and therapy, 10(3), 365–
386. https://doi.org/10.1007/s13555-020-00372-0.
DermNet. (2021). Introduction to Dermoscopy.
https://dermnetnz.org/cme/dermoscopy-course/introdu
ction-to-dermoscopy/.
FastAI. (2021). https://docs.fast.ai/.
Fu’adah, Y.N., Caecar Pratiwi1, NK., Adnan Pramudito1,
M., and Nur Ibrahim, N. (2020). Convolutional Neural
Network for Automatic Skin Cancer Classification
System. doi:10.1088/1757-899X/982/1/012005.
Gholamiangonabadi, D., Kiselov, N., & Grolinger, K.
(2020). Deep Neural Networks for Human Activity
Recognition With Wearable Sensors: Leave-One-
Subject-Out Cross-Validation for Model Selection.
IEEE Access. doi: 10.1109/ACCESS.2020.3010715.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep
Residual Learning for Image Recognition. The IEEE
Conference on Computer Vision and Pattern
Recognition (CVPR). doi: 10.1109/CVPR.2016.90.
ImageNet. (2021). https://www.image-net.org/index.php.
ISIC. (2018). Skin Lesion Analysis Towards Melanoma
Detection. https://challenge2018.isic-archive.com/.
ISIC. (2019). Skin Lesion Analysis Towards Melanoma
Detection. https://challenge2019.isic-archive.com/.
Krizhevsky, A. (2009). https://www.cs.toronto.edu/~kriz/
cifar.html.
Le, T.T. & Nguyen, H.Q. (2017). Automatic Skin Lesion
Analysis Towards Melanoma Detection. The 21
st
Asia
Pacific Symposium on Intelligent and Evolutionary
Systems. doi:10.1109/IESYS.2017.8233570.
Mahbod, A., Schaefer, G., Wang, C., Ecker, R., & I.
Ellinge, I. (2019). Skin Lesion Classification Using
Hybrid Deep Neural Networks. The IEEE International
Conference on Acoustics, Speech and Signal
Processing (ICASSP). doi: 10.1109/ICASSP.2019.86
83352.
Nasr-Esfahani, E., et al. (2016). Melanoma Detection by
Analysis of Clinical Images Using Convolutional
Neural Network. The 38
th
Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society (EMBC). doi: 10.1109/EMBC.2016.7
590963.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image
Segmentation. The 18th International Conference on
Medical Image Computing and Computer Assisted
Intervention. doi:10.1007/978-3-319-24574-4_28.
Rundo, F., Banna, G.L., Conoci, S. (2019). Bio-Inspired
Deep-CNN Pipeline for Skin Cancer Early Diagnosis.
Computation 2019, 7, 44. https://doi.org/10.3390/
computation7030044.
Sanjay, M. (2018). Why and how to Cross Validate a
Model?. https://towardsdatascience.com/why-and-
how-to-cross-validate-a-model-d6424b45261f.
Shaban, M. (2020). Deep Convolutional Neural Network
for Parkinson’s Disease Based Handwriting Screening.
The IEEE 17
th
International Symposium on Biomedical
Imaging Workshops. doi: 10.1109/ISBIWorkshops502
23.2020.9153407.
Shi, W., et al. (2016). Real-Time Single Image and Video
Super-Resolution Using an Efficient Sub-Pixel
Convolutional Neural Network. The IEEE Conference
on Computer Vision and Pattern Recognition.
doi:10.1109/CVPR.2016.207.
Simonyan, K. and A. Zisserman, A. (2015). Very Deep
Convolutional Networks for Large-Scale Image
Recognition. The International Conference on Learning
Representations.
Stefan Jianu, S.R., et al. (2019). Automatic Diagnosis of
Skin Cancer Using Neural Networks. The 11
th
International Symposium on Advanced Topics in
Electrical Engineering. doi: 10.1109/ATEE.2019.87
24938.
Sun, Y., Xue, B., Zhang, M. and Yen, G.G. (2020).
Completely Automated CNN Architecture Design
Based on Blocks. The IEEE Transactions on Neural
Networks and Learning Systems, vol. 31, no. 4, pp.
1242-1254. doi: 10.1109/TNNLS.2019.2919608.
Szegedy, C., et al. (2015). Going Deeper with
Convolutions. The IEEE Conference on Computer
Vision and Pattern Recognition.
Worcester Polytechnic Institute (WPI). (2018). High
Performance Computing. https://arc.wpi.edu/
computing/hpc-clusters/.
AutoCNN-MSCD: An Autodesigned CNN Framework for Detecting Multi-skin Cancer Diseases over Dermoscopic Images
615
