
Multi Modality Medical Image Translation for Dicom Brain Images
Ninad Anklesaria
1
, Yashvi Malu
1
, Dhyey Nikalwala
1
, Urmi Pathak
1
, Jinal Patel
1
,
Nirali Nanavati
1
, Preethi Srinivasan
2
and Arnav Bhavsar
2
1
Department of Computer Engineering, Sarvajanik College of Engineering & Technology, Surat, India
2
School of Computing and Electrical Engineering, Indian Institute of Technology Mandi, Mandi, India
Keywords:
MRI, T1 Weighted-image Modality, T2 Weighted-Image Modality, Image Translation, DICOM, U-Net.
Abstract:
The acquisition time for different MRI (Magnetic Resonance Imaging) image modalities pose a unique chal-
lenge to the efficient usage of the contemporary radiology technologies. The ability to synthesize one modality
from another can benefit the diagnostic utility of the scans. Currently, all the exploration in the field of medi-
cal image to image translation is focused on NIfTI (Neuroimaging Informatics Technology Initiative) images.
However, DICOM (Bidgood et al., 1997) images are the prevalent image standard in MRI centers. Here,
we propose a modified deep learning network based on U-Net architecture for T1-Weighted image (T1WI)
modality to T2-Weighted image (T2WI) modality image to image translation for DICOM images and vice
versa. Our deep learning model exploits the pixel wise features between T1W images and T2W images which
are important to understand the brain structures. The observations indicate better performance of our approach
to the previous state-of-the-art methods. Our approach can help to decrease the acquisition time required for
the scans and thus, also avoid motion artifacts.
1 INTRODUCTION
Medical imaging allows us to see the processes going
on inside our body without the need of surgery or any
invasive procedure. MRI is one type of medical imag-
ing technique used to create diagnostic images with-
out the use of any harmful radiation. Different MRI
sequences are used to optimize tissue contrast and in-
crease the diversity of diagnostic information. The
various MRI sequences are as follows: T1WI, T2WI,
FLAIR (Fluid attenuated inversion recovery), Proton
density, Diffusion weighted, STIR (Short Tau Inver-
sion Recovery). MRI images are generated by vary-
ing TR (repetition time) and TE (Echo time) times in
MRI machines (Preston, 2006).
The most commonly referenced MRI sequences
for diagnosis are T1WI and T2WI. In T1WI images
CSF (Cerebrospinal fluid) and inflammation (infec-
tion, demyelination) is dark, white matter is light,
cortex is gray and fat (within the bone marrow) is
bright.In T2WI images. CSF and inflammation (in-
fection, demyelination) is bright, white matter is dark
gray, cortex is light grey and fat (within the bone mar-
row) is bright (Preston, 2006).
Although clinical judgement may be sufficient for
prognosis of many conditions, use of medical imaging
for diagnostics helps in confirming, correctly assess-
ing and documenting courses of many diseases and in
assessing responses to treatment. The sequential ac-
quisition of medical images is time consuming for the
radiologist and costly for the patient. Long periods of
data collection are a major source of motion artifacts
which contributes to inferior quality of scans. Short-
ening of acquisition time could potentially induce a
more cost and time efficient system for both the pa-
tients and radiologists.
From a brief survey at Johns Hopkins and some
surrounding MRI practices in Baltimore suggest that
routine imaging times for a wide range of examina-
tions vary from 20 to 60 minutes (Edelstein et al.,
2010). Each protocol frequently includes 5 or more
pulse sequences (Edelstein et al., 2010). As quoted
by the radiologists we consulted, T1 Weighted-Image
takes ≈ 5 minutes to generate and T2-Weighted-
Image takes ≈ 6-7 minutes. The proposed solution
takes ≈ 14.3 seconds to generate T2WI from T1WI
using an NVIDIA Tesla K80 9 GB.
For an MRI image, k space has to be built. K-
space refers to the data matrix that contains raw MRI
data obtained directly from the MRI scanner before
Fourier Transformation is applied to get the final im-
age. With the progress in deep learning and image
processing, a technique called image to image trans-
lation is developed that can help to reduce the acqui-
sition time. The aim is to transfer one type of image
to another while preserving the content.
168
Anklesaria, N., Malu, Y., Nikalwala, D., Pathak, U., Patel, J., Nanavati, N., Srinivasan, P. and Bhavsar, A.
Multi Modality Medical Image Translation for Dicom Brain Images.
DOI: 10.5220/0010906400003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 168-173
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

With the development of neural networks, this
task is able to be unified into a single problem: given
pairs of example images from both domains, teach a
convolutional neural network to map the input images
to the output images. Use of image to image transla-
tion in medical imaging is to generate images virtu-
ally, images which are not acquired due to the clinical
workflow.
Various neural networks have been developed and
used for image to image translation. Generative ad-
versarial network (GAN) is the most popular model
used for the same. By performing various transfor-
mations on the basic GAN model, various other net-
works viz. cGAN (Isola et al., 2016), Pix2Pix (Isola
et al., 2016), MedGAN (Armanious et al., 2018), Cy-
cleGAN (Zhu et al., 2017) have been developed.
In this paper, we propose a multimodality (T1WI
to T2WI and vice versa) image translation model for
DICOM brain images. DICOM images are the preva-
lent medical industry standard and DICOM images
are smaller in size compared to the corresponding
NIfTI images. We discuss the pre-processing tech-
niques for DICOM data as well as the proposed U-Net
based model in this paper. We further show a qualita-
tive and quantitative comparison of the generated re-
sults with the ground truth followed by the scope of
future work.
2 RELATED WORK
Numerous contributions have been made in literature
on medical image translation. However, most of these
contributions pertain to the NIfTI format which are
used for research purposes. A deep network based so-
lution to reconstruct T2WI from T1WI and few sam-
ples of k-space for T2WI using an encoder-decoder
architecture has been proposed on NIfTI brain images
in (Srinivasan et al., 2020). A comparison for image-
to-image translation of T1WI and T2WI is proposed
using CycleGAN and U-Net for NIfTI brain images
in (Welander et al., 2018). Considering the impor-
tance of complementary information present in differ-
ent modalities and the predominant industrial usage,
DICOM images are for the first time, motivated to be
utilized in construction of T2WI from a given T1WI
using our proposed U-Net based model.
The advantages and uses of image-to-image trans-
lation on paired and unpaired images using GANs es-
pecially in medical imaging using deep learning has
been explained in (Kaji and Kida, 2019) (Alotaibi,
2020) (Shen et al., 2019). In (Avula, 2020), Convo-
lutional Neural Network (CNNs) specialising in vi-
sual imagery are explored for the reconstruction of T1
Weighted Glioma Images from T1 Weighted-Images.
Conditional Generative Adversarial Net-
works(cGAN) which enables fine-tuned contrast
synthesis are tested in (Yang et al., 2020) for cross
modality registration and MRI segmentation to
perform cross modality image-to-image translation
of MRI scans. Predictive Generative Adversarial
Networks(pGAN) method is compared with cGAN in
(Dar et al., 2018) where both utilize adversarial loss
functions and correlated structure across neighboring
cross-sections for improved synthesis, particularly
at high spatial frequencies. In (Xiao et al., 2018),
the authors demonstrate an algorithm that learns
complex mappings between different MRI contrasts
and accurately transforms between T1WI and T2WI,
proton density images, time-of-flight angiograms,
and diffusion MRI images. A tool to transform non
T1W-Images to have a similar contrast profile to
an adult T1W-Image as mentioned in (Neurabenn,
2020) has been developed which uses the basic
U-Net model. Whole medical image synthesis using
Deep Encoder-Decoder Image Synthesizer has been
proposed in (Sevetlidis et al., 2016).
3 PROPOSED WORK
Several kind of deep learning models were investi-
gated during the literature study. Two models stood
out amongst the others in synthesizing realistic im-
ages in high resolution - Encoder-decoder and U-Net.
In medical image to image translation, paired images
from source and target modality are needed. Conver-
sion of one modality to another modality uses extrac-
tion of features like tissues and fat cells.
U-Net (Ronneberger et al., 2015) can be consid-
ered as a modified version of encoder decoder archi-
tecture. As shown in Figure 2, U-Net architecture
consists of a contracting path to capture context and a
symmetric expanding path that enables precise local-
ization. The main idea is to supplement a contracting
network by successive layers, where pooling opera-
tors are replaced by up-sampling operators. These
layers increase the resolution of the output. A suc-
cessive convolution layer can then learn to assemble
a more precise output based on this information.
U-Net architecture is divided into 2 parts – a con-
tracting path and an expansive part. As we can see in
Figure 2, in the contracting path, the spatial dimen-
sions are reduced and the number of channels are in-
creased while in the expansive path, dimensions are
increased and channels are decreased. Then, with a
set of transformations, we end up with high-resolution
features which are then combined to predict a relevant
Multi Modality Medical Image Translation for Dicom Brain Images
169

target value from our images. Our proposed U-Net
based model reduces over-fitting and produces com-
petitive output for T1WI to T2WI images and vice
versa.
3.1 Proposed Model for T1WI to T2WI
Translation
A python package called Pydicom is used as the MRI
images are in DICOM format. Firstly, the images are
read using read file and then converted into numpy ar-
rays using pixel array function. From this numpy ar-
ray, we calculate the x-gradient and y-gradient using
the Sobel function. The x- and y-gradient help in bet-
ter edge detection of the brain structure. So, 3 numpy
arrays - original image, x-gradient and y-gradient are
used to train the neural network to learn the semantic
transformation for the required mapping of T1WI to
T2WI (Srinivasan et al., 2020).
The contracting path of our proposed model con-
sists of repeated application of two 3x3 convolutions
with a dropout layer between them followed by a 2x2
max pooling operation for down-sampling. The drop-
out layers help reduce over-fitting in our proposed
model.
The number of feature channels are doubled after
each step. The expansive path consists of upsampling
of feature channels using a 2x2 conv2DTranspose
(up-convolution) followed by concatenation between
the output of the up-convolution with the feature map
from the contracting path. This is followed by two
3x3 convolutions with a dropout layer between them.
At the final layer a 1x1 convolution is used to map
the feature vector to the desired number of classes
(Neurabenn, 2020).
The output from the neural network which is the
numpy array containing the generated image is then
converted into .dcm(DICOM) format using PixelData
function.
4 DATASET AND EVALUATION
METRICS
The dataset acquired in order to carry out the evalu-
ation consisted of paired T1W and T2W images for
21 subjects. Per subject 18 axial slices were available
from segregated brain scans. The data was split into a
training set of 90 slices and a testing set of 36 slices.
The accuracy for the simulated image was calcu-
lated on the basis of 3 metrics: PSNR (Peak Signal to
Noise Ratio), MSE (Mean Squared Error) and MAE
(Mean Absolute Error) as given in Equation 1, 2 and
3. As the MRI images usually differ in intensity, ev-
ery image is normalized using min-max normaliza-
tion. As the intensity values obtained from the pixel
array are of varying scales they tend to contribute un-
equally in model fitting and model learning function
which in turn results in making the model biased. So
we use min max normalization to deal with this prob-
lem.
As an entire new image is being generated the ac-
curacy can be calculated on the basis of loss it experi-
ences. Loss function is used to evaluate how much the
synthetic images are different from the ground truth.
The metrics can provide a theoretical assurance for
the quality of the image.
Mean Squared Error (MSE):
MSE =
1
n
n
∑
i=1
(y
i
− ˜y
i
)
2
(1)
where y
i
is the numpy array of T2 ground truth and
˜y
i
is the numpy array of generated T2.
Peak Signal-To-Noise Ratio (PSNR) :
PSNR = 10 ∗ log
10
MAX
2
1
MSE
(2)
where MAX
I
is the maximum possible pixel value of
the image
Mean Absolute Error (MAE):
MAE =
1
n
n
∑
i=1
|y
i
− x
i
| (3)
where y
i
is the numpy array of T2 ground truth and
x
i
is the numpy array of generated T2.
5 RESULTS AND ANALYSIS
5.1 Qualitative Comparison
We consulted radiologist in our city for their feedback
on our results and their remark was that the simulated
DICOM images have graphically naturalistic presen-
tation. T2WI were prone to being misclassified, due
to the fact that the sombre (dark) appearance of T2W
images can lead to confusion regarding the authentic-
ity of noise which could be present in T2WI.
In Figure 1 and Figure 5, we show the recon-
structed T2WI vis-a-vis the ground truth. The con-
tinuous nature of the edges of simulated T2WI in Fig-
ure 1 could be explained due to the fact that each brain
BIOIMAGING 2022 - 9th International Conference on Bioimaging
170
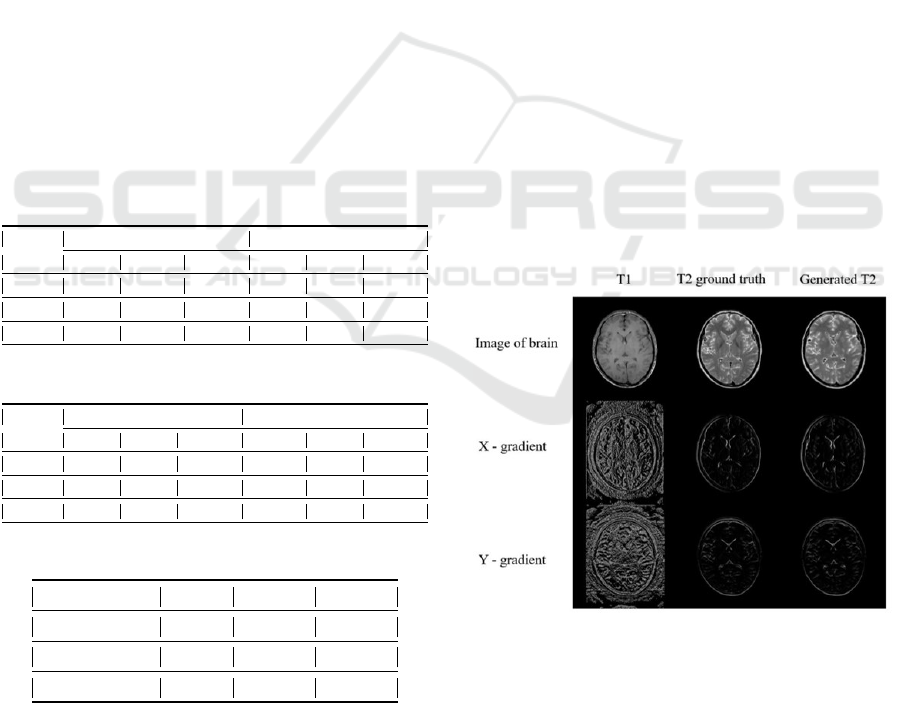
image has a distinctive shape, and CSF(Cerebrospinal
fluid) and fat tissues(within bone marrow) are bright
in T2WI. The portion of images where the intensity
varies significantly i.e. CSF and white matter, appear
to be confusing for the model to learn, this also could
be a result of disparity between contrasting images of
different subjects. Further, in Figure 3 and Figure 4
we have shown the reconstructed T1WI and shown a
visual comparison with the ground truth.
In the U-Net model, during the convolution, the
perceptual detail of the image is diminished while
feature detail is improved. The expansive pathway
of the U-Net combines the feature and perceptual
detail through a progression of up-convolutions and
concatenations with high-resolution features from the
convulsing path. If during the up-convolutions the
aim is set to be minimizing the error difference of
simulated images from the ground truth, the simulated
images may be one step ahead.
5.2 Quantitative Comparison
Quantitative results from the Encoder-Decoder archi-
tecture, our proposed U-Net architecture and Unet
(Ronneberger et al., 2015) are shown in the Table 1,
Table 2 and Table 3.
Table 1: Metric result of T1WI to T2WI.
No. of
Epochs
Encoder-Decoder Proposed Unet Architecture
MSE MAE PSNR MSE MAE PSNR
150 0.0236 0.0859 16.2769 0.0006 0.0073 31.9393
200 0.0167 0.07296 17.7756 0.0006 0.0065 32.510
250 0.0156 0.07212 18.0581 0.0005 0.0063 32.7199
Table 2: Metric result of T2WI to T1WI.
No. of
Epochs
Encoder-Decoder Proposed Unet Architecture
MSE MAE PSNR MSE MAE PSNR
150 0.0381 0.1227 14.1889 0.0009 0.0109 30.4947
200 0.0313 0.1072 15.0511 0.00017 0.0188 27.7326
250 0.0345 0.1122 14.6216 0.0007 0.0091 31.6396
Table 3: Metric result of T1WI to T2W2I using Unet.
No. of Epochs MSE MAE PSNR
150 0.012 0.10011 19.2019
200 0.0035 0.0224 24.556
250 0.0076 0.0628 21.1683
The U-Net architecture outperforms the Encoder-
Decoder model in all quantitative measurements for
T1 images to T2 images and vice versa. Further, our
proposed modified UNET architecture outperforms
the Unet (Ronneberger et al., 2015) architecture.
6 FUTURE SCOPE
A suggestion for future work is to investigate other
modalities of MRI like FLAIR, Proton density,
diffusion-MRI, etc on DICOM dataset. Another pos-
sible aspect is to work with different body organs like
cervical spine, prostate gland, liver, kidney, bladders,
etc. Here we have implemented a DICOM dataset on
U-Net and encoder decoder (Srinivasan et al., 2020)
architecture. The same can be applied on various
known and forthcoming architectures which might
enhance the results with low memory usage.
The application of Image to Image translation on
DICOM can further be realized in modalities of var-
ious medical imaging techniques most notably, CT
scan, PET scan, X-rays.
7 SUMMARY
The proposed architecture is based on the U-Net ar-
chitecture and the use of x-gradient and y-gradient
lead to better reconstruction results of DICOM im-
ages and can be utilized for other similar reconstruc-
tion.
8 ILLUSTRATION
Figure 1: Reconstructed T2WI from T1WI (.jpg format).
Multi Modality Medical Image Translation for Dicom Brain Images
171
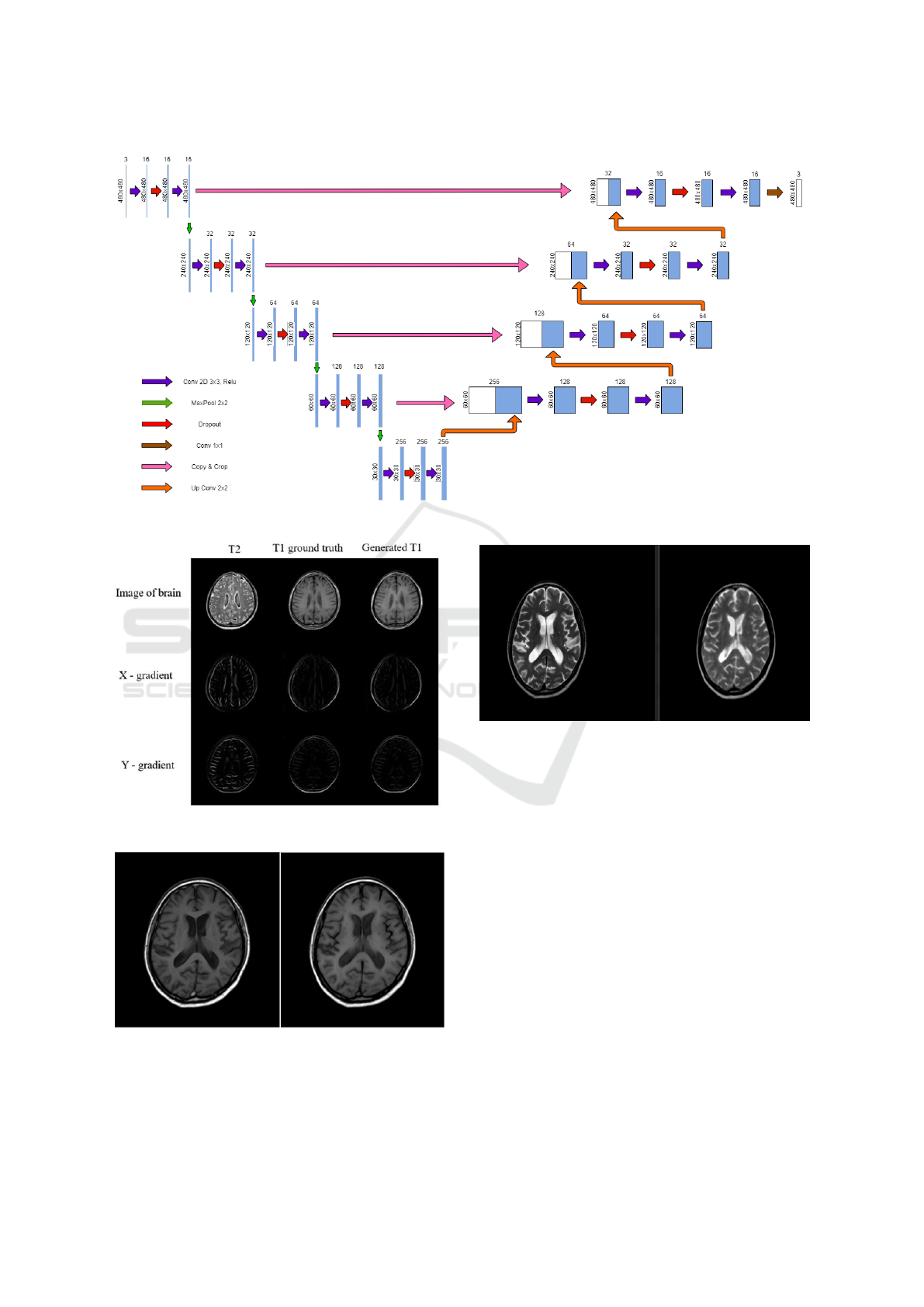
Figure 2: Proposed Unet Architecture.
Figure 3: Reconstructed T1WI from T2WI (.jpg format).
Figure 4: Reconstructed T1WI in DICOM format (ground
truth vs reconstructed).
Figure 5: Reconstructed T2WI in DICOM format.
ACKNOWLEDGEMENTS
We are thankful for the MRI data (DICOM brain im-
ages in different modalities) provided to us by Dr.
Nipun Jindal from Superscan Imaging Centre, Surat
solely for research purpose. This was really instru-
mental in our research work.
REFERENCES
Alotaibi, A. (2020). Deep generative adversarial networks
for image-to-image translation: A review. volume 12.
Armanious, K., Yang, C., Fischer, M., K
¨
ustner, T., Niko-
laou, K., Gatidis, S., and Yang, B. (2018). Medgan:
Medical image translation using gans. volume
abs/1806.06397.
Avula, A. (2020). Medical image translation using convo-
lutional neural networks.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
172

Bidgood, Jr, W. D., Horii, S. C., Prior, F. W., and
Van Syckle, D. E. (1997). Understanding and using
DICOM, the data interchange standard for biomedical
imaging. volume 4, pages 199–212. Oxford Univer-
sity Press (OUP).
Dar, S. U. H., Yurt, M., Karacan, L., Erdem, A., Erdem,
E., and C¸ ukur, T. (2018). Image synthesis in multi-
contrast mri with conditional generative adversarial
networks.
Edelstein, W. A., Mahesh, M., and Carrino, J. A. (2010).
Mri: time is dose–and money and versatility. vol-
ume 7, pages 650–652. 20678736[pmid].
Isola, P., Zhu, J., Zhou, T., and Efros, A. A. (2016). Image-
to-image translation with conditional adversarial net-
works. volume abs/1611.07004.
Kaji, S. and Kida, S. (2019). Overview of image-to-image
translation by use of deep neural networks: denois-
ing, super-resolution, modality conversion, and recon-
struction in medical imaging.
Neurabenn (2020). Neurabenn/t1ify: Applying style trans-
fer to convert multi-modal mri images to have a t1 like
contrast.
Preston, D. C. (2006). Magnetic resonance imaging (mri)
of the brain and spine: Basics.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation.
Sevetlidis, V., Giuffrida, V., and Tsaftaris, S. (2016). Whole
image synthesis using a deep encoder-decoder net-
work. pages 127–137.
Shen, Z., Huang, M., Shi, J., Xue, X., and Huang, T. S.
(2019). Towards instance-level image-to-image trans-
lation. IEEE Computer Society.
Srinivasan, P., Kaur, P., Nigam, A., and Bhavsar, A. (2020).
Semantic features aided multi-scale reconstruction of
inter-modality magnetic resonance images.
Welander, P., Karlsson, S., and Eklund, A. (2018). Gener-
ative adversarial networks for image-to-image trans-
lation on multi-contrast mr images - a comparison of
cyclegan and unit.
Xiao, S., Wu, Y., Lee, A. Y., and Rokem, A. (2018).
Mri2mri: A deep convolutional network that accu-
rately transforms between brain mri contrasts. Cold
Spring Harbor Laboratory.
Yang, Q., Li, N., Zhao, Z., Fan, X., Chang, E. I.-C., and Xu,
Y. (2020). Mri cross-modality image-to-image trans-
lation. volume 10, page 3753.
Zhu, J., Park, T., Isola, P., and Efros, A. A. (2017). Unpaired
image-to-image translation using cycle-consistent ad-
versarial networks. volume abs/1703.10593.
Multi Modality Medical Image Translation for Dicom Brain Images
173
