
Semantic Segmentation of Retinal Blood Vessels from Fundus Images by
using CNN and the Random Forest Algorithm
Ayoub Skouta
1 a
, Abdelali Elmoufidi
2 b
, Said Jai-Andaloussi
1 c
and Ouail Ouchetto
1 d
1
Computer and Systems Laboratory, Hassan II University, Casablanca, Morocco
2
Data4Earth Laboratory, Sultan Moulay Slimane University, Beni Mellal, Morocco
{ay.skouta, elmoufidi10, andaloussi.said, ouail.ouchetto}@gmail.com
Keywords:
Funds Images, Diabetic Retinopathy, CAD System, Semantic Segmentation, Blood Vessel Detection,
Artificial Intelligence, Deep Learning, Convolutional Neural Networks.
Abstract:
Abstract: In this paper, we present a new study to improve the automated segmentation of blood vessels in
diabetic retinopathy images. Pre-processing is necessary due to the contrast between the blood vessels and the
background, as well as the uneven illumination of the retinal images, in order to produce better quality data to
be used in further processing. We use data augmentation techniques to increase the amount of accessible data
in the dataset to overcome the data sparsity problem that deep learning requires. We then use the CNN VGG16
architecture to extract the feature from the preprocessed background images. The Random Forest method will
then use the extracted attributes as input parameters. We used part of the augmented dataset to train the model
(1764 images, representing the training set); the rest of the dataset will be used to test the model (196 images,
representing the test set). Regarding the model validation phase, we used the dedicated part for testing the
DRIVE dataset. Promising results compared to the state of the art were obtained. The method achieved an
accuracy of 98.7%, a sensitivity of 97.4% and specificity of 99.5%. A comparison with some recent previous
work in the literature has shown a significant advancement in our proposal.
1 INTRODUCTION
The human eye is a visual organ, similar to a sphere
with a diameter of 2-3 cm, weighing about 8 grams. It
receives the light emitted by objects, focuses it, then
transmits the information to the brain for analysis.
The eye is surrounded by a tough membrane called
the sclera. The main structures of the eyeball are :
The cornea is completely transparent and is located
in front of the eye in front of the iris, but because it
is transparent, it cannot be seen. It can be observed
with a special microscope called a slit lamp. When
we look at an eye, we can see that the iris is colored
and shrinks depending on the light, and that the sclera
is white and opaque and covers the entire back sur-
face of the eye. The crystalline lens is a converging
lens-like organ that reflects the image and directs it
to the retina. The figure shows a human eye with all
its parts (Abr
`
amoff et al., 2010). The retina is the in-
a
https://orcid.org/0000-0002-6176-910X
b
https://orcid.org/0000-0002-8574-9584
c
https://orcid.org/0000-0002-6864-1141
d
https://orcid.org/0000-0001-8287-215X
ner membrane of the eyeball. It is a light sensitive
layer that captures light rays. The retina is made up
of three layers of nerve cells: ganglion cells, bipo-
lar cells and visual cells that are either cones or rods.
Light passes through the upper layers of the retina
to reach the cones and rods. After a series of pho-
tochemical reactions, the information is transmitted
to the bipolar cells, then to the ganglion cells and fi-
nally to the optic nerves, which send information to
the brain in the form of electrical signals, which in
turn interpret the signals received as visual images.
The cones are grouped in the center of the retina at
the macula, where visual acuity is best. They are the
color vision cells that work only in well-lit environ-
ments. The rods appear around the macula, they are
the cells of black and white vision they react to very
weak lights. Progressive damage to the macula leads
to diseases such as macular degeneration or, in severe
cases, creates a macular hole, which causes blood ves-
sels in the macula to rupture (Jakobiec, 1982). Dia-
betes is a comprehensive metabolic disorder that can
lead to various vascular complications in the body.
There are two types of diabetes: type 1 diabetes and
type 2 diabetes. Both result in high blood sugar lev-
Skouta, A., Elmoufidi, A., Jai-Andaloussi, S. and Ouchetto, O.
Semantic Segmentation of Retinal Blood Vessels from Fundus Images by using CNN and the Random Forest Algorithm.
DOI: 10.5220/0010911800003118
In Proceedings of the 11th International Conference on Sensor Networks (SENSORNETS 2022), pages 163-170
ISBN: 978-989-758-551-7; ISSN: 2184-4380
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
163
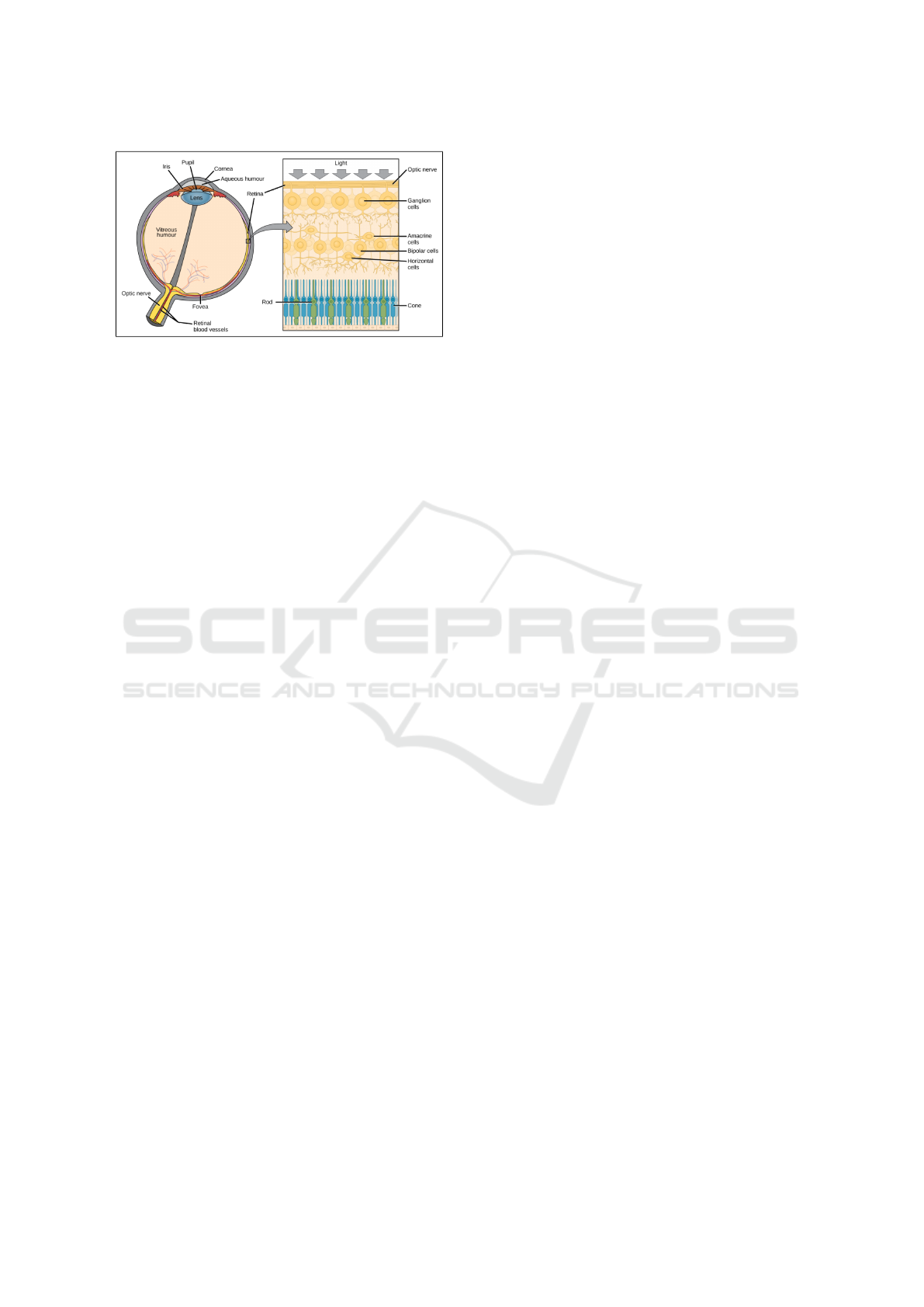
Figure 1: This anatomy of the eye and the retina.
els related to a hormone called insulin produced by
the pancreas; this hormone is responsible for the reg-
ulation of blood sugar levels in the blood (Breiman,
2001a). Diabetic retinopathy is a microangiopathy
that has the manifestations of occlusion and leakage
of microvascular fluid and blood in the retina, it is es-
sential to understand the signs of occlusion and leak-
age in the retina before understanding the pathogen-
esis and sign of diabetic retinopathy. In the presence
of diabetes and hyperglycemia, several things happen
in the blood vessels, the blood vessel walls that help
nourish the retina deteriorate, the blood cells become
distorted and the blood thickens and then finally a mi-
crovascular occlusion as there will be irregularities in
the blood flow and a decrease in oxygen. This re-
sults in visual manifestations called lesions such as
microaneurysms, hemorrhages, hard exudates, cot-
tony spots (Agurto et al., 2010). This phenomenon
due to diabetes can progressively damage the struc-
ture of the eyeball, leading to severe vision loss or
even blindness (Kanski and Bowling, 2011). Several
studies have focused on the use of automated tech-
niques for the early diagnosis of diabetic retinopa-
thy. These technologies are becoming increasingly
advantageous in the healthcare sector, thanks to ar-
tificial intelligence (AI), which allows the creation
of reliable software for decision support in the diag-
nosis of medical images (Tham Chen2014g, ). The
use of traditional machine learning (ML) approaches
such as random forest algorithm, decision tree, near-
est neighbor method (KNN), K-means, support vector
method (SVM), etc. (Kotsiantis et al., 2007), (Liaw
et al., 2002) are used to analyze fundus images to de-
velop models that can predict outcomes without pro-
gramming, through operations distributed over mul-
tiple layers (Kapoor et al., 2019). Machine learn-
ing techniques are divided into two categories: su-
pervised and unsupervised learning. The supervised
technique requires labeled data to train the segmenta-
tion model, but the unsupervised method requires no
prior knowledge of the labeled data to train the seg-
mentation model. Since the introduction of convo-
lutional neural networks (CNNs), deep learning has
been remarkably successful, and it is now considered
the most successful AI model in all computer vision
applications. It has proven to be very effective and
useful in extracting features for use in various ma-
chine learning applications. The input layer, hidden
layers and output layer are the three types of layers
found in CNNs. These layers consist of linked nodes,
each with its own output layer sending a weighted
quantity to the activation function. Several strategies
have been established to improve the quality of vas-
cular segmentation, using classical machine learning
or deep learning methods, but more can be done. The
second section of this paper contains an overview of
related previous work, and then the proposed method
and materials used are described in the third section
of this study. The results of our experiment are de-
scribed in Section 4. A discussion is presented in Sec-
tion Five. Finally, the main conclusions are presented
in the last section.
2 RELATED PREVIOUS WORKS
In the medical domain, many computer-aided diag-
noses have used to help in diagnosing of many de-
ceases. Even more, many diseases are early-detected
using artificial intelligence. And one of the newest
examples is the creation of two models one to clas-
sify a COVID-19 test of a suspect patient as posi-
tive or negative, and the other to classify the hospital-
ization units of patients with COVID-19 (de Oliveira
et al., 2021). Others diseases are breast cancer (El-
moufidi et al., 2018), (Elmoufidi et al., 2014), (Elm-
oufidi et al., 2015), (Elmoufidi, 2019) and diabetic
retinopathy (Skouta et al., 2021), (El Hossi et al.,
2021), (Stabingis et al., 2018), (Balkys and Dzemyda,
2012). In this paper, the purpose of determining reti-
nal vascular segmentation is to predict abnormali-
ties that can occur in the retina, including diabetic
retinopathy and glaucoma. The advent of convolu-
tional neural networks has led to improvements in re-
ducing the burden on specialists with screening pro-
grams that have begun to generate high-accuracy seg-
mentation (Salazar-Gonzalez et al., 2014). Accord-
ing to the literature presented in this section, different
methods are applied to segment blood vessels in fun-
dus images. In this paper, we focus on the use of deep
CNNs, which have shown excellent performance in
their application. The remainder of this section will
be devoted to presenting related work on blood vessel
segmentation from retinal fundus images; for exam-
ple, the approach proposed by Khalaf et al (Khalaf
SENSORNETS 2022 - 11th International Conference on Sensor Networks
164
et al., 2016) used a CNN to segment vessels, using an
input patch derived from the original image as input.
Three convolutional layers and a fully connected layer
at the end of the network constitute the CNN. Their
technique classifies each patch into three categories
based on the center pixel: background, large vessels,
and small vessels. Liskowski and Krawiec (Liskowski
and Krawiec, 2016) presented another patch-based
approach to determine the number of neurons in the fi-
nal fully connected layer. Background and vessels are
the two categories predicted by CNN. Segmentation
is performed by connecting each pixel to a neuron, so
that the CNN output is a two-dimensional vector. Yu
et al (Yu et al., 2020) developed a CNN for hierar-
chical vessel division by first extracting vessel trees
using a graph-based method and then classifying the
retinal vasculature using two algorithms (PLDA and
AHCA). The final layers of the CNN are fully con-
nected layers. Shuangling Wang et al (Wang et al.,
2015) designed a retinal blood vessel segmentation
technique that combines a CNN as a feature extractor
from raw images and a Random Forest (RF) algorithm
as a classifier. Zhou et al (Zhou et al., 2017) propose
a CNN solution to extract blood vessel features. The
extracted features are then provided to the dense CRF
to perform segmentation. The DeepVessel CNN pub-
lished by Fu et al. (Fu et al., 2016) combines the out-
put side layer and the CRF layer to model long-range
pixel interactions and nonlocal pixel correlations. Hu
et al (Hu et al., 2018) wrote a paper that performs
binary segmentation by describing an RCF-inspired
multi-scale CNN architecture that provides a com-
prehensive description of vascular features combined
with an improved cross-entropy loss function. The
emergence of the U-net architecture in 2015 (Ron-
neberger et al., 2015) has made a great contribution in
the field of biomedical image segmentation. It has be-
come popular in blood vessel segmentation. Several
authors have customized this promising method to de-
velop modified versions; Zhang and Chung (Zhang
and Chung, 2018), Yan et al. (Yan et al., 2018), Yan
et al. (Yang et al., 2017), Wu et al. (Wu et al., 2018),
Wu et al. (Wu et al., 2020a), Wang et al. (Wang et al.,
2020), Wu et al. (Wu et al., 2019), Wang et al. (Wang
et al., 2019), Ma et al. (Ma et al., 2019), Mishra et al.
(Mishra et al., 2020).
2.1 Materials
In this section, we outline the hardware used, the
datasets used and the proposed methodology. We
tested our solution in a GPU environment using the
Keras library and TensorFlow.
2.2 Databases Used
The suggested approach has been trained, validated
and tested using the Drive, HRF datasets. These
datasets are freely accessible and are used in the
vast majority of vascular segmentation studies. The
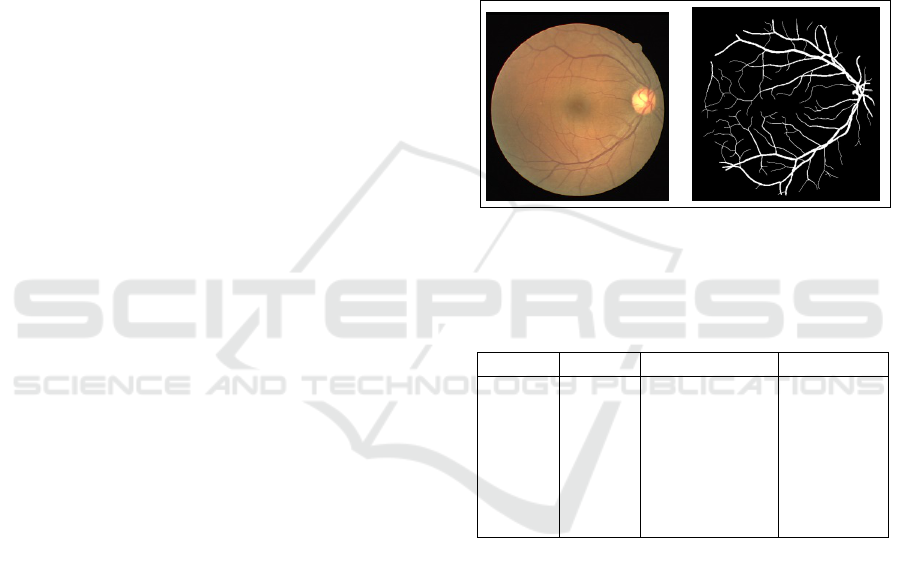
DRIVE (Digital Retinal Image for Vessel Extraction)
dataset is a series of 20 images that includes a manual
segmentation of vessels for each image. There are 20
more photos in the test set. The images have a size of
768 x 584 pixels. the exception of seven photos that
showed slight symptomatic RD, all images DRIVE
were normal (Staal et al., 2004).
Figure 2: Left: Examples of fundus from the DERIVE
database. Right: Examples of ground truth data from the
DERIVE database.
Table 1: Presentation of the databases implemented in the
proposed approach.
Datasets Source Images Digitizer
Drive
Avail-
able:
Online
Staal
et al.
(Wu
et al.,
2019)
40 (33
healthy, 7
mild early
DR) Res-
olutions :
768x584
Canon: CR5
non-
mydriatic
3CCD
fundus
camera,
FOV 45
2.3 Image Preprocessing
The high similarity in the fundus image between the
vessels and the background can lead to erroneous seg-
mentation. The data is optimized in the preprocessing
step to provide a better image with a clear distinc-
tion between the vessels and the background, helping
CNN to interpret the input images and allows for cor-
rect segmentation. This step starts by cropping the
black border of the original images. After the orig-
inal color image is cropped, the color order is BGR
(blue, green, red) instead of RGB, so the first step
is to convert the image from BGR to RGB. We then
extract the green channel from the multichannel im-
age after splitting it into several channels, which gives
more contrast between the blood vessels and the back-
Semantic Segmentation of Retinal Blood Vessels from Fundus Images by using CNN and the Random Forest Algorithm
165
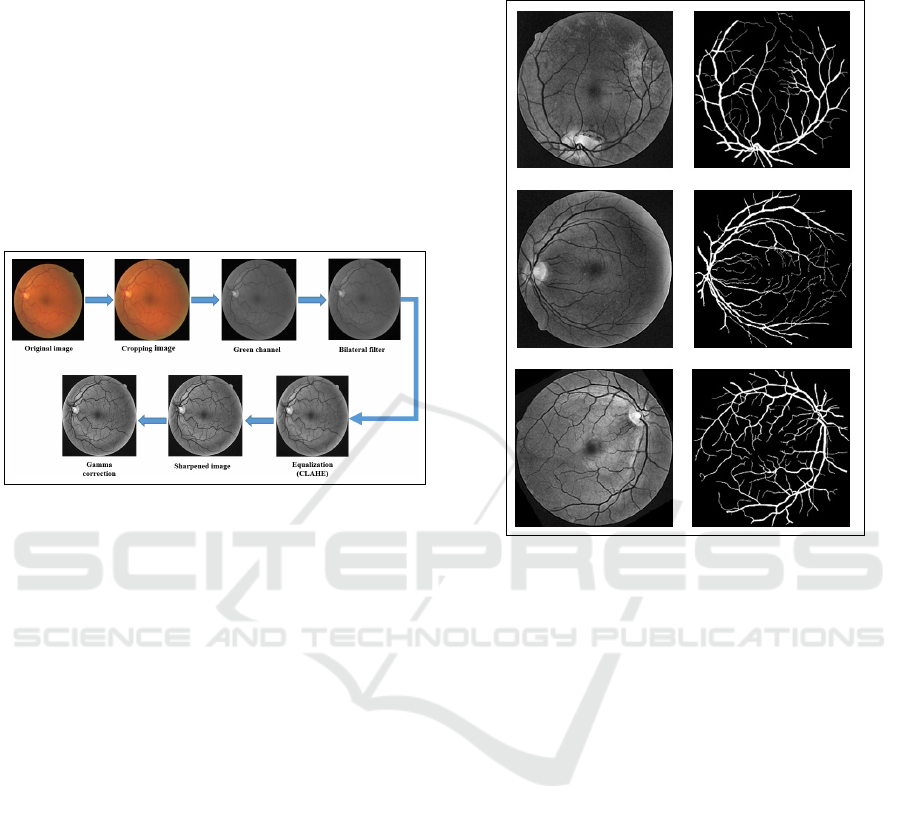
ground than the red and blue channels. The green
channel is then smoothed and noise is reduced us-
ing a bilateral filter while vascular contours are pre-
served. Vascular information is further highlighted by
applying contrast-limited adaptive histogram equal-
ization (CLAHE) to the image generated by the bi-
lateral filter. We used the image sharpening method
to highlight each individual pixel, enhance the color
it emanates, and increase the pixel density, making it
more visible to CNN. Finally, a gamma correction, to
correct the retinal image in order to remove the un-
even light factor. The results of the pre-processing
are shown in Figure 3.
Figure 3: Result of preprocessing step.
2.4 Data Augmentation
The step of data augmentation is proposed of this
study; it consists in modifying the existing pre-
processed images in the original database as well as
its associated mask in order to increase the size of the
dataset that will be used for the training and testing
of the proposed method. Data augmentation strate-
gies reduce overfitting and give the proposed model
more power while also enhancing accuracy and re-
silience. Having a CNN with a huge dataset increases
the model’s capacity to generalize to new pictures
from other databases. To achieve this, we employ
a number of techniques to generate various kinds of
sample while preserving the attributes of the source
images: The original images and the images obtained
after the preprocessing stage, together with their ac-
companying ground-truth masks, are rotated from 0
to 360
◦
at a 30
◦
angle each time. Randomly, we add
noise, change the brightness, change the colorimetry,
vertical and horizontal flips and horizontal and verti-
cal flips to each image created by the loop that rotates
the image from 0 to 360 degrees. These are the trans-
formations used in the data update process. The total
data recovered from the original photos and the im-
ages acquired after the pre-processing step is equal
to 1960 images, which are divided into 1764 images,
representing the training set (90 percent of the gener-
ated images) and 196 images, representing the vali-
dation set (ten percent of the total generated images).
Figure 4: An example of images after the data augmentation
procedure.
3 PROPOSED METHODOLOGY
3.1 Feature Extraction by CNN
Segmentation is the classification of pixels, where
each pixel, rather than the whole picture, is assigned
to a distinct category. The feature extraction approach
is critical for a successful segmentation process. Ex-
traction is a critical operation because the features
used to define the candidate areas have a direct impact
on the accuracy with which each pixel in the input
image is classified as a blood vessel or background.
The CNN has recently demonstrated the capacity to
recognize the most sophisticated, basic, and signif-
icant visual elements such as edges, corners, orien-
tated edges, and so on. This is crucial information
for the analysis and categorization of pixels. Follow-
ing that, subsequent layers integrate these character-
istics to capture higher order features (Wang et al.,
2019). In this technique, the CNN VGG16 architec-
ture is utilized as a feature extractor for pixels in terms
of quantifiable metrics that can be used in the clas-
sification stage to determine if the pixels correspond
to real blood from the vessel or not. The Random
SENSORNETS 2022 - 11th International Conference on Sensor Networks
166

Forest classifier (Breiman, 2001b) takes the role of
the VGG16 model’s last fully connected layer. This
method is known as transfer learning, and it involves
merely training the top level of the network. The
VGG16 architecture is composed of 5 blocks with a
total of 16 layers. Essentially it is based on the use of
3x3 convolution with stride equal to 1 and a ”Same”
padding, i.e. the size of the input image matches
the size of the output image. After each convolution
layer, a max 2x2 pooling layer is used to reduce the
size.
3.1.1 Segmentation using the Random Forest
Algorithm
Our goal is to take pretrained VGG16 weights used as
feature extractors and then segment the retinal images
using a random drill. The images resulting from the
data augmentation operation are physically stored on
the hard disk of our computer, and then we have to
load all the images and their associated masks. Since
CNN is suitable for large images, we kept the origi-
nal size of the images in our training database, which
have a height of 584 and width of 565, and then they
are converted from RGB to BGR because the opencv
library simply reads the images as BGR. Next, we im-
ported the VGG16 model, so now we will also import
the weights corresponding to the weights in the im-
agenet database. The include top variable is set to
false, this basically means that we have not imported
the dense layers and the output layer. The size of the
input images of the VGG16 model is changed as the
model defaults to 244x244x3 at the size of our input
images. The next step is to apply the feature extrac-
tor to our training data and see what the features look
like. As shown in Figure 5, the features of the origi-
nal augmented images and the preprocessed and aug-
mented images are merged into a bag of features. The
Figure 5: Displays of extracted characteristics.
number of features generated is glaring, therefore we
have eliminated features such as the features that rep-
resent the background of the retinal image. These fea-
tures are useless to use in training, as they will require
additional training time. The pixels that represent the
blood vessels are kept as features. After the feature
reduction operation we feed the random forest algo-
rithm to segment the retinal blood vessels.
4 EXPERIMENTAL RESULT
4.1 Evaluation Metrics
We examine the performance of the segmentation
findings with relation to the expert’s manual segmen-
tation using the following performance measures: ac-
curacy, sensitivity, and specificity as indicators.
Table 2: Presentation of the databases implemented in the
proposed approach.
Metrics Formule Description
Sensitivity
T P
T P + FN
The ratio of correctly cat-
egorized vascular pixels
compared to genuine vas-
cular pixels is known as
the true positive rate.
Specificity
T N
T N + FP
As compared to real non-
vascular pixels, the frac-
tion of correctly diag-
nosed non-vascular pix-
els.
Accuracy
T N + T P
T P + FP + TN + FN
As a proportion of the to-
tal number of pixels in the
picture, the accuracy rep-
resents how well blood
• True positives (TP) refers to the number of accu-
rately segmented blood vessel pixels;
• The amount of accurately divided background
pixels is shown by True Negatives (TN);
• False positives (FP) are background pixels that
have been segmented incorrectly into blood ves-
sel pixels;
• False negatives (FN) are pixels in blood vessels
that have been mistakenly labeled background.
4.2 Results
We used the DRIVE dataset to train the suggested
models in 100 epochs. The results of our method’s
segmentation are more substantial. The accuracy is
98.7%, the sensitivity is 97.4%, and the specificity
is 99.5 percent. Training details are depicted in the
Semantic Segmentation of Retinal Blood Vessels from Fundus Images by using CNN and the Random Forest Algorithm
167
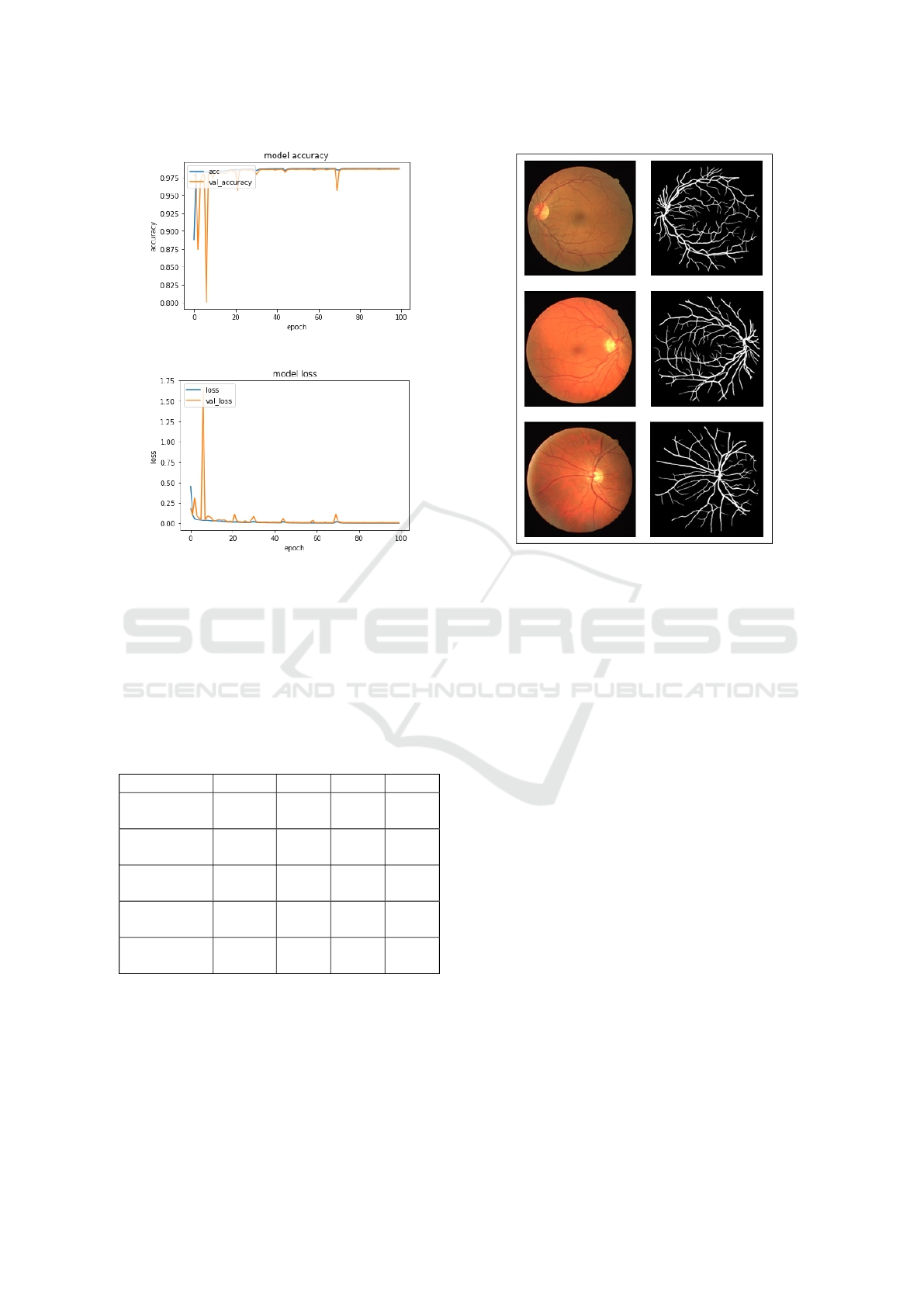
Figure 6: Model accuracy performance.
Figure 7: Model loss performance.
figures, which also show the accuracy and losses at-
tained.
We used a subset of the DRIVE database dedi-
cated to testing. Our primary aim is to verify the accu-
racy of our suggested technique. We examine the de-
gree of similarity between the segmentations obtained
using our proposed network and the ground truth.
Table 3: Comparison of segmentation performances for
DRIVE.
Works Year Acc Sen Spe
(Tamim
et al., 2020)
2020 0.9607 0.7542 0.9843
(Tian et al.,
2020)
2020 0.958 0.8639 0.9690
(Wu et al.,
2020b)
2020 0.9582 0.7996 0.9813
(Boudegga
et al., 2021)
2021 0.9819 0.8448 0.99
Proposed
method
2021 98.7 97.4 99.5
Acc: Accuracy Sen: Sensitivity Spe: Specificity
5 CONCLUSIONS
Early treatment of diabetic retinopathy with blood
vessel segmentation helps people with diabetes avoid
severe visual loss. Deep learning is one of the most
Figure 8: Comparisons of segmentation results using the
test subset of the DRIVE database.
sophisticated methods for segmentation challenges,
as it improves accuracy. The effective convolutional
neural network architecture used will help ophthal-
mologists to eradicate vision loss related to diabetic
retinopathy. In this research, we suggest the use of a
VGG16 model to extract characteristics and combine
it with the random forest technique to automate blood
vessel segmentation. Our technique was developed
from the DRIVE dataset, which has been shown to be
resilient.
REFERENCES
Michael D Abr
`
amoff, Mona K Garvin, and Milan Sonka.
Retinal imaging and image analysis. IEEE reviews in
biomedical engineering, 3:169–208, 2010.
Frederick A Jakobiec. Ocular anatomy, embryology, and
teratology. Harpercollins, 1982.
Leo Breiman. Random forest, vol. 45. Mach Learn, 1,
2001a.
Carla Agurto, Victor Murray, Eduardo Barriga, Sergio
Murillo, Marios Pattichis, Herbert Davis, Stephen
Russell, Michael Abr
`
amoff, and Peter Soliz. Multi-
scale am-fm methods for diabetic retinopathy lesion
detection. IEEE transactions on medical imaging, 29
(2):502–512, 2010.
Jack J Kanski and Brad Bowling. Clinical ophthalmol-
ogy: a systematic approach. Elsevier Health Sciences,
2011.
SENSORNETS 2022 - 11th International Conference on Sensor Networks
168

Sotiris B Kotsiantis, I Zaharakis, P Pintelas, et al. Su-
pervised machine learning: A review of classification
techniques. Emerging artificial intelligence applica-
tions in computer engineering, 160(1):3–24, 2007.
Andy Liaw, Matthew Wiener, et al. Classification and re-
gression by randomforest. R news, 2(3):18–22, 2002.
Rahul Kapoor, Stephen P Walters, and Lama A Al-Aswad.
The current state of artificial intelligence in ophthal-
mology. Survey of ophthalmology, 64(2):233–240,
2019.
Rodrigo Felipe Albuquerque Paiva de Oliveira, Carmelo
Jose Albanez Bastos Filho, Ana Clara AMVF
de Medeiros, Pedro Jose Buarque Lins dos Santos,
and Daniela Lopes Freire. Machine learning applied
in sars-cov-2 covid 19 screening using clinical analy-
sis parameters. IEEE Latin America Transactions, 19
(6):978–985, 2021.
Abdelali Elmoufidi, Khalid El Fahssi, Said Jai-Andaloussi,
Abderrahim Sekkaki, Quellec Gwenole, and Mathieu
Lamard. Anomaly classification in digital mammog-
raphy based on multiple-instance learning. IET Image
Processing, 12(3):320–328, 2018.
Abdelali Elmoufidi, Khalid El Fahssi, Said Jai-Andaloussi,
Nabil Madrane, and Abderrahim Sekkaki. Detection
of regions of interest’s in mammograms by using lo-
cal binary pattern, dynamic k-means algorithm and
gray level co-occurrence matrix. In 2014 Interna-
tional Conference on Next Generation Networks and
Services (NGNS), pages 118–123. IEEE, 2014.
Abdelali Elmoufidi, Khalid El Fahssi, Said Jai-Andaloussi,
and Abderrahim Sekkaki. Automatically density
based breast segmentation for mammograms by us-
ing dynamic k-means algorithm and seed based re-
gion growing. In 2015 IEEE International Instru-
mentation and Measurement Technology Conference
(I2MTC) Proceedings, pages 533–538. IEEE, 2015.
Abdelali Elmoufidi. Pre-processing algorithms on digital x-
ray mammograms. In 2019 IEEE International Smart
Cities Conference (ISC2), pages 87–92. IEEE, 2019.
Ayoub Skouta, Abdelali Elmoufidi, Said Jai-Andaloussi,
and Ouail Ochetto. Automated binary classification
of diabetic retinopathy by convolutional neural net-
works. In Advances on Smart and Soft Computing,
pages 177–187. Springer, 2021.
Amine El Hossi, Ayoub Skouta, Abdelali Elmoufidi, and
Mourad Nachaoui. Applied cnn for automatic diabetic
retinopathy assessment using fundus images. In Inter-
national Conference on Business Intelligence, pages
425–433. Springer, 2021.
Giedrius Stabingis, Jolita Bernatavi
ˇ
cien
˙
e, Gintautas Dze-
myda, Alvydas Paunksnis, Lijana Stabingien
˙
e, Povi-
las Treigys, and Ramut
˙
e Vai
ˇ
caitien
˙
e. Adaptive eye
fundus vessel classification for automatic artery and
vein diameter ratio evaluation. Informatica, 29(4):
757–771, 2018.
Gediminas Balkys and Gintautas Dzemyda. Segmenting the
eye fundus images for identification of blood vessels.
Mathematical Modelling and Analysis, 17(1):21–30,
2012.
Ana Salazar-Gonzalez, Djibril Kaba, Yongmin Li, and Xi-
aohui Liu. Segmentation of the blood vessels and op-
tic disk in retinal images. IEEE journal of biomedical
and health informatics, 18(6):1874–1886, 2014.
Aya F Khalaf, Inas A Yassine, and Ahmed S Fahmy. Con-
volutional neural networks for deep feature learning
in retinal vessel segmentation. In 2016 IEEE Interna-
tional Conference on Image Processing (ICIP), pages
385–388. IEEE, 2016.
Paweł Liskowski and Krzysztof Krawiec. Segmenting reti-
nal blood vessels with deep neural networks. IEEE
transactions on medical imaging, 35(11):2369–2380,
2016.
Linfang Yu, Zhen Qin, Tianming Zhuang, Yi Ding,
Zhiguang Qin, and Kim-Kwang Raymond Choo. A
framework for hierarchical division of retinal vascular
networks. Neurocomputing, 392:221–232, 2020.
Shuangling Wang, Yilong Yin, Guibao Cao, Benzheng Wei,
Yuanjie Zheng, and Gongping Yang. Hierarchical reti-
nal blood vessel segmentation based on feature and
ensemble learning. Neurocomputing, 149:708–717,
2015.
Lei Zhou, Qi Yu, Xun Xu, Yun Gu, and Jie Yang. Improv-
ing dense conditional random field for retinal vessel
segmentation by discriminative feature learning and
thin-vessel enhancement. Computer methods and pro-
grams in biomedicine, 148:13–25, 2017.
Huazhu Fu, Yanwu Xu, Stephen Lin, Damon Wing Kee
Wong, and Jiang Liu. Deepvessel: Retinal vessel seg-
mentation via deep learning and conditional random
field. In International conference on medical image
computing and computer-assisted intervention, pages
132–139. Springer, 2016.
Kai Hu, Zhenzhen Zhang, Xiaorui Niu, Yuan Zhang, Chun-
hong Cao, Fen Xiao, and Xieping Gao. Retinal vessel
segmentation of color fundus images using multiscale
convolutional neural network with an improved cross-
entropy loss function. Neurocomputing, 309:179–191,
2018.
Olaf Ronneberger, Philipp Fischer, and Thomas Brox. U-
net: Convolutional networks for biomedical image
segmentation. In International Conference on Medi-
cal image computing and computer-assisted interven-
tion, pages 234–241. Springer, 2015.
Yishuo Zhang and Albert CS Chung. Deep supervision with
additional labels for retinal vessel segmentation task.
In International conference on medical image comput-
ing and computer-assisted intervention, pages 83–91.
Springer, 2018.
Zengqiang Yan, Xin Yang, and Kwang-Ting Cheng. A
three-stage deep learning model for accurate retinal
vessel segmentation. IEEE journal of Biomedical and
Health Informatics, 23(4):1427–1436, 2018.
Yehui Yang, Tao Li, Wensi Li, Haishan Wu, Wei Fan, and
Wensheng Zhang. Lesion detection and grading of di-
abetic retinopathy via two-stages deep convolutional
neural networks. In International Conference on Med-
ical Image Computing and Computer-Assisted Inter-
vention, pages 533–540. Springer, 2017.
Yicheng Wu, Yong Xia, Yang Song, Yanning Zhang, and
Weidong Cai. Multiscale network followed network
model for retinal vessel segmentation. In Inter-
national Conference on Medical Image Computing
Semantic Segmentation of Retinal Blood Vessels from Fundus Images by using CNN and the Random Forest Algorithm
169

and Computer-Assisted Intervention, pages 119–126.
Springer, 2018.
Yicheng Wu, Yong Xia, Yang Song, Yanning Zhang, and
Weidong Cai. Nfn+: A novel network followed net-
work for retinal vessel segmentation. Neural Net-
works, 126:153–162, 2020a.
Kun Wang, Xiaohong Zhang, Sheng Huang, Qiuli Wang,
and Feiyu Chen. Ctf-net: Retinal vessel segmenta-
tion via deep coarse-to-fine supervision network. In
2020 IEEE 17th International Symposium on Biomed-
ical Imaging (ISBI), pages 1237–1241. IEEE, 2020.
Yicheng Wu, Yong Xia, Yang Song, Donghao Zhang,
Dongnan Liu, Chaoyi Zhang, and Weidong Cai.
Vessel-net: retinal vessel segmentation under multi-
path supervision. In International Conference on
Medical Image Computing and Computer-Assisted In-
tervention, pages 264–272. Springer, 2019.
Bo Wang, Shuang Qiu, and Huiguang He. Dual encod-
ing u-net for retinal vessel segmentation. In In-
ternational Conference on Medical Image Comput-
ing and Computer-Assisted Intervention, pages 84–
92. Springer, 2019.
Wenao Ma, Shuang Yu, Kai Ma, Jiexiang Wang, Xinghao
Ding, and Yefeng Zheng. Multi-task neural networks
with spatial activation for retinal vessel segmentation
and artery/vein classification. In International Con-
ference on Medical Image Computing and Computer-
Assisted Intervention, pages 769–778. Springer, 2019.
Suraj Mishra, Danny Z Chen, and X Sharon Hu. A data-
aware deep supervised method for retinal vessel seg-
mentation. In 2020 IEEE 17th International Sym-
posium on Biomedical Imaging (ISBI), pages 1254–
1257. IEEE, 2020.
Joes Staal, Michael D Abr
`
amoff, Meindert Niemeijer,
Max A Viergever, and Bram Van Ginneken. Ridge-
based vessel segmentation in color images of the
retina. IEEE transactions on medical imaging, 23(4):
501–509, 2004.
Leo Breiman. Random forest, vol. 45. Mach Learn, 1,
2001b.
Nasser Tamim, Mohamed Elshrkawey, Gamil Abdel Azim,
and Hamed Nassar. Retinal blood vessel segmenta-
tion using hybrid features and multi-layer perceptron
neural networks. Symmetry, 12(6):894, 2020.
Chun Tian, Tao Fang, Yingle Fan, and Wei Wu. Multi-path
convolutional neural network in fundus segmentation
of blood vessels. Biocybernetics and Biomedical En-
gineering, 40(2):583–595, 2020.
Yicheng Wu, Yong Xia, Yang Song, Yanning Zhang, and
Weidong Cai. Nfn+: A novel network followed net-
work for retinal vessel segmentation. Neural Net-
works, 126:153–162, 2020b.
Henda Boudegga, Yaroub Elloumi, Mohamed Akil,
Mohamed Hedi Bedoui, Rostom Kachouri, and
Asma Ben Abdallah. Fast and efficient retinal blood
vessel segmentation method based on deep learning
network. Computerized Medical Imaging and Graph-
ics, 90:101902, 2021.
SENSORNETS 2022 - 11th International Conference on Sensor Networks
170
