
Replicability of Differentially Expressed Genes Versus Biological
Pathways Biomarkers in Diagnosing Sepsis
Kelsey Winkeler
1 a
and Carly A. Bobak
2 b
1
Department of Psychology, Virginia Polytechnic Institute and State University Blacksburg, VA 24060, U.S.A.
2
Biomedical Data Science, Dartmouth College, Hanover, NH 03755, U.S.A.
Keywords:
Differentially Expressed Genes, Sepsis, Replicability, Biological Pathways.
Abstract:
It is generally believed that biological pathways representing curated gene sets are not only more interpretable,
but also more replicable and reproducible than gene signatures. With the falling costs of next generation se-
quencing, we are approaching a point where the cost fully sequencing the transcriptome is competitive with
quantifying a targeted gene expression signature which opens up the possibility of pathway signatures for in-
fectious disease. In this work, we evaluated if pathway based signatures are really more reproducible than gene
signatures (improvement between 0.83 and over 1 million fold), and amend a meta-analysis framework known
for generating highly reproducible gene signatures to instead produce pathway signatures (AUC improves
from 0.854 to 0.964 and 0.556 to 0.677 between gene and pathway signatures in independent validation data).
We conclude that pathway based signatures show clinical promise for the diagnosis of infectious disease, and
there is a growing need for methods considering such signatures.
1 INTRODUCTION
Reproducibility and replicability, wherein discrimi-
natory biological features are consistently associated
with a phenotype both within the same dataset and
across new datasets, is a major challenge to using
differentially expressed genes (DEGs) for diagnoses
(Crow et al., 2019; Sweeney et al., 2015). Most ap-
proaches take minimal biological information into ac-
count, struggle to remain consistent across samples
and platforms, and make data difficult to interpret bi-
ologically. (Tan, 2003; Zhang et al., 2008) Recent
efforts have been made to combine gene expression
with knowledge of biological pathways and function
using Gene Ontology (GO) terms and other annotated
pathways. These methods have the advantage of be-
ing less complex and more biologically interpretable
than traditional analysis of DEGs, emphasizing net-
works of related genes over individual genes. (Khatri
et al., 2012; Zhang et al., 2009)
There has been some successes: Zarringhalam et
al. predicted kidney transplant rejection and response
to Infliximab in ulcerative colitis (Zarringhalam et al.,
2014), and Pradines et al. found improved repro-
a
https://orcid.org/0000-0002-9452-9428
b
https://orcid.org/0000-0001-8631-4753
ducibility within and between datasets in several dis-
eases (Pradines et al., 2020). Such approached may
be useful even in studies with small sample sizes.
(Lim et al., 2015) While prior attempts at diagnostics
aim to reduce biomarker quantity to make any result-
ing assay more cost-effective to produce, the falling
cost of transcriptome-wide sequencing makes these
proposed pathway signatures possible.(Alpern et al.,
2019; Mayday et al., 2019; Sholder et al., 2020)
Sepsis is a particularly important potential appli-
cation, as it can present similarly to the non-infectious
systemic inflammatory response syndrome (SIRS)
and lacks a rapid, gold-standard diagnostic. Begin-
ning treatment within the first ‘golden hour’ is integral
for reducing mortality in severe sepsis; however, inap-
propriate and overuse of antibiotics in hospitals con-
tinues to increase rates of MRSA and other antibiotic-
resistant microbes. (Ferrer et al., 2014; van Zanten,
2014; Cohen et al., 2015; Sweeney and Khatri, 2017)
Attempts to create a sepsis diagnostic DEG signa-
ture include the sepsis meta score (Sweeney et al.,
2015), FA1M3:PLAC8 ratio (Scicluna et al., 2015),
and the SeptiCyte lab (McHugh et al., 2015), the three
of which were compared using 39 public data sets
in Sweeney and Khatri in 2017(Sweeney and Khatri,
2017) and demonstrated a similar ability to discrimi-
nate between patients with and without sepsis.
160
Winkeler, K. and Bobak, C.
Replicability of Differentially Expressed Genes Versus Biological Pathways Biomarkers in Diagnosing Sepsis.
DOI: 10.5220/0010976300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 3: BIOINFORMATICS, pages 160-167
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

In this work, we extend the current research com-
paring pathway biomarkers to DEGs by specifically
focusing on the implications of pathway biomarkers
in diagnosis pipelines. Specifically, we consider com-
paring the overall rank of important DEGs to the rank
of important pathways across multiple datasets col-
lected on multiple gene expression array platforms,
and evaluate how a pathway based approach could be
amended to a meta-analysis framework to further im-
prove replicability of diagnostic biomarkers.
2 METHODS
All analyses were performed in R version 4.0.3. (R
Core Team, 2017)
2.1 Data Sources
Following Sweeney et al.(Sweeney and Khatri, 2017),
we downloaded publicly available human gene ex-
pression microarray datasets from the NIH Gene
Expression Omnibus (GEO). (Barrett et al., 2012;
Sutherland et al., 2011; Dolinay et al., 2012; Par-
nell et al., 2012; McHugh et al., 2015; Kim et al.,
2021) Cases included blood samples from adult pa-
tients with severe infection or sepsis, taken on day
1 of presentation and pre-treatment. Controls were
limited to hospitalization patients without infectious
diagnoses. A summary of the included datasets is
shown in Table 1. Of the discovered datasets, two
were withheld for diagnostic validation while three
were used for discovery purposes.
2.2 Processing and Biomarker Analysis
Normalized, publicly available datasets were
downloaded using the ‘MetaIntegrator’ package in
R.(Haynes et al., 2016) Probe IDs were matched to
HUGO gene symbol; where multiple probes matched
to the same symbol, the median expression value was
used. (Tweedie et al., 2021) Differentially expressed
genes (DEGs) were identified using the ‘limma’
package in R. (Ritchie et al., 2015)
We conducted pathway analysis using two meth-
ods. First, we conducted Gene Set Enrichment Anal-
ysis (GSEA) using the results from the DEGs de-
scribed above. (Mootha et al., 2003; Subrama-
nian et al., 2005) Genes were first ranked using
sign( f oldchange) × (− log
10
(p-value)) as described
previously (Chen et al., 2007). Ranked lists were im-
ported into the GSEA 4.1.0 and enriched or depleted
pathways were identified from Gene Ontology: Bi-
ological Processes (GO:BP),(Ashburner et al., 2000)
Reactome,(Wu and Haw, 2017) BioCyc Genome
Database Collection (Karp et al., 2019) and Wikipath-
ways(Martens et al., 2021). Only pathways between
5 and 500 genes were considered.
Second, we used single sample Gene Set Enrich-
ment Analysis (ssGSEA) to reduce the dimension of
each gene expression matrix to a pathway expression
matrix using the ‘GSVA’ package in R (H
¨
anzelmann
et al., 2013). Following above, we only mapped to
pathways between 5 and 500 genes. We then repeated
the ‘limma’ analysis described to identify DEGs using
pathways as biomarker signatures in lieu of genes.
2.3 Replicability of Ranked Lists
The results from the DEGs, GSEA, and ssGSEA
were ranked according in each discovery dataset. To
compare ranked lists, we used Rank Biased Overlap
(RBO). RBO has the benefit of being appropriate for
ranked lists, particularly lists which may contain dif-
ferent numbers of elements and is often used in com-
paring search results (Webber et al., 2010).
2.4 Meta Analysis and Diagnostic Score
Following Sweeney et al. (Sweeney et al., 2015;
Sweeney and Khatri, 2017), we used the meta integra-
tor package (Haynes et al., 2016) on the gene expres-
sion matrices and ssGSEA pathway matrices to con-
duct a random effects meta analysis of possible sepsis
biomarkers in the discovery datasets. Gene/pathway
biomarkers that had a summary FDR p-value < 0.01,
heterogeneity p-value < 0.05, or were not signifi-
cant in all three discovery datasets were filtered out
from further analysis. We first considered the ‘top’
biomarker results by taking the 10 biomarkers with
the highest magnitude in summary effect size. We
also constructed a diagnostic score using the greedy
forward based search defined in Sweeney et al.,
wherein the biomarker with the most discriminatory
power is added first, and then subsequent biomark-
ers are added based on their improvement to area un-
der the receiving operator characteristics (AUROC)
curves until AUROC no longer improves. This score
is used to construct AUROC curves in both the dis-
covery and validation datasets to compare how gene
or pathway biomarker signatures perform in discrimi-
nating sepsis cases across multiple datasets (Sweeney
et al., 2015; Sweeney and Khatri, 2017; Haynes et al.,
2016).
Replicability of Differentially Expressed Genes Versus Biological Pathways Biomarkers in Diagnosing Sepsis
161
Table 1: The experimental design for the datasets included in the reproducibility analysis.
Split Control definition Case Definition n Sample
GSE28750 Validation 24h after ’major surgery’ community-acquired sepsis 21 Blood
GSE32707 Discovery MICU with or without
SIRS, nonseptic
Sepsis, sepsis/ARDS 103 Blood
GSE40012 Discovery SIRS Sepsis from community ac-
quired pneumonia
31 Blood
GSE74224 Discovery Post-surgical infection-
negative systemic inflam-
mation
Sepsis patients from ICU 105 Blood
GSE66099 Validation SIRS Sepsis, Septic Shock 229 Blood
Table 2: The RBO results from comparing ranked DEG and pathways from GSEA or ssGSEA.
DEG GSEA ssGSEA FC GSEA FC ssGSEA
GSE32707 vs GSE40012 7.53E-08 4.88E-02 5.45E-07 648 142.48 6.24
GSE32707 vs GSE74224 5.73E-05 1.05E-04 1.99E-04 0.83 2.47
GSE40012 vs GSE74224 7.94E-08 5.43E-02 8.89E-02 683 966.30 1 119 232.86
3 RESULTS AND DISCUSSION
We identified 5 datasets containing blood samples
from adult patients with sepsis or severe infection.
Three of these datasets were selected for biomarker
discovery, and two were withheld for validation.
Table 1 describes the experimental design of each
dataset. Cases were considered to be sepsis or se-
vere infection, and controls were post-surgical pa-
tients, patients with trauma, and or systemic inflam-
matory response syndrome (SIRS).
Complete ranked lists of DEGs were compared
between the three discovery cohorts. The RBO for
each comparison is reported in Table 2. Median RBO
across the three comparisons was 7.94 × 10
−08
. Path-
ways identified using GSEA were then ranked by
and compared using the RBO metric. The RBO for
the GSEA analysis is also reported in Table 2. Me-
dian RBO across the three GSEA comparisons was
4.88 × 10
−02
. To illustrate the improvement in RBO
similarity, we calculated the fold change improve-
ment in RBO between DEG and GSEA identified
pathways. As shown in Table 2, the improvement was
large, with two comparisons having a fold change im-
provement of over 6 × 10
5
.
While it is unsurprising that ranked lists of
pathways are more reproducible than ranked genes,
canonical methods like GSEA consider pathways at a
dataset level rather than an individual level. Hence,
we should consider whether sample level pathway
analysis is more reproducible than DEGs using the
same analysis techniques that would typically be used
to identify transcriptomic biomarkers.
ssGSEA uses gene expression data to identify a
pathway score for each sample in a data matrix. The
median RBO across our comparisons for ‘differen-
tially expressed’ pathways (DEPs) is 1.99 × 10
−04
,
with each comparison shown in Table 2. Similar to
the GSEA pathways, all comparisons had consider-
able improvement compared to the DEG ranking.
Despite this improvement in reproducibility, the
RBO was low across both ranked pathways and
ranked genes, reflective of biological complexity. It is
also unclear how these improvements in reproducibil-
ity will affect diagnostic performance at the clinical
level. To study this further, we used the meta-analysis
model proposed by Sweeney et al. (Sweeney et al.,
2015; Sweeney and Khatri, 2017) to compare the re-
sults between the gene and pathway signatures.
A critical step of this method is filtering biomark-
ers based on summary effects size, significance, and
heterogeneity (Cochran’s Q) (Sweeney et al., 2015;
Sweeney and Khatri, 2017). While filters based on
the summary effects yielded a similar number of
biomarkers, filtering based on heterogeneity removed
far more genes than pathways. The heterogeneity step
reduced the gene biomarker list to 48 positively and
6 negatively associated genes, conversely, the hetero-
geneity filter reduced the pathway biomarker list to 55
positive and 50 negative pathways.
The meta analysis results of top 10 associated
genes, by magnitude of effect size, is shown in Figure
1a while the corresponding top pathways are shown in
Figure 1b. The distributions of the summary estimates
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
162
Table 3: The genes selected by the greedy forward search in a meta-analysis framework for the diagnosis of sepsis.
Gene FC p-value BH p-value previously associated with sepsis
ADORA2A 1.033 1.63E-06 5.21E-04 Yes (Busse et al., 2016)
ARSD 0.863 9.56E-10 1.64E-06 Yes (Guillen-Guio et al., 2020)
LY6G6D 0.646 3.12E-06 7.78E-04 No
SIGLEC9 0.982 6.14E-12 5.70E-08 Yes (von Gunten et al., 2009)
ZBTB7B 0.648 2.87E-06 7.41E-04 Yes (Bhatty et al., 2012)
TTC17 0.651 2.59E-06 6.96E-04 No
GADD45G 0.643 3.47E-06 8.15E-04 Yes (Aare et al., 2012)
PPP1R9B
0.644 3.32E-06 8.02E-04 No
PARP10 0.634 4.72E-06 9.94E-04 Yes (Wasyluk and Zwolak, 2021)
MPZL2 -0.862 9.69E-10 1.64E-06 Yes (Ji et al., 2014)
DLG5 -0.710 3.36E-07 1.59E-04 Yes (Li et al., 2017)
EPHB4 -0.647 3.14E-06 7.78E-04 Yes (Coulthard et al., 2012)
Table 4: The pathways selected by the greedy forward search in a meta-analysis framework for the diagnosis of sepsis.
Name ID FC p-value BH p-value gene count
Negative regulation of myeloid cell
differentiation
GO:0045638 0.996 3.23E-12 2.82E-08 59
Cell killing GO:0001906 0.723 2.14E-07 6.67E-05 27
Negative regulation of tumor necro-
sis factor superfamily cytokine pro-
duction
GO:1903556 0.955 3.13E-07 8.64E-05 29
Regulation of fatty acid metabolic
process
GO:0019217 -0.698 8.28E-06 7.72E-04 62
APEX1-Independent resolution of
AP sites via the single nucleotide re-
placment pathway
R-HSA-5649702 -0.695 1.26E-05 9.95E-04 7
Polyamine biosyntethic process GO:0006596 -0.624 6.31E-06 6.47E-04 9
Signalling by PTK6 R-HSA-8848021 -0.650 2.72E-06 3.41E-04 61
Regulation of cell cycle G1/S phase
transition
GO:1902806 -0.781 2.45E-08 2.33E-05 110
between the most significant genes and pathways is
similar (Kolmogorov-Smirnov p=0.1641), suggesting
that biomarker signatures should be competitive be-
tween genes and pathways.
We identified a reduced biomarker signature in
both gene expression and pathway matrices using a
greedy forward search algorithm, where biomarkers
were selected based on maximizing the AUC between
cases and controls in the discovery datasets. The fi-
nal meta-score gene signature for sepsis genes can be
found in Table 3 and the meta-score pathway signa-
ture in Table 4.
AUROC for the discovery datasets were promis-
ing, using either the gene meta-score or pathway
meta-score. The AUROCs using the gene meta-
score are 0.815, 0.925, and 0.952 for GSE32707,
GSE40012, and GSE74224 respectively. The AU-
ROCs using the pathway meta-score are 0.833, 0.884,
and 0.925 in GSE32707, GSE40012, and GSE74224
respectively. AUROCs were not significantly differ-
ent between the gene signature and the pathway sig-
nature in the discovery datasets.
The AUROC for the validation datasets is shown
in Figure 2. Considerable improvement is seen in the
pathway signature in both datasets. Of note, both
signatures perform poorly in GSE66099, which is
an amalgamation of 6 unique sepsis datasets and in-
cludes patients with septic shock. In particular, the
gene based signature performs only slightly better
than random guessing while the pathway based sig-
nature exhibits poor performance with an AUROC of
0.667. Despite the improvement in a pathway based
signature, more work is needed to increase repro-
ducibility and replicability in validation datasets be-
fore such a signature would be considered for use in a
clinic. However, the improvement of AUROC in the
Replicability of Differentially Expressed Genes Versus Biological Pathways Biomarkers in Diagnosing Sepsis
163
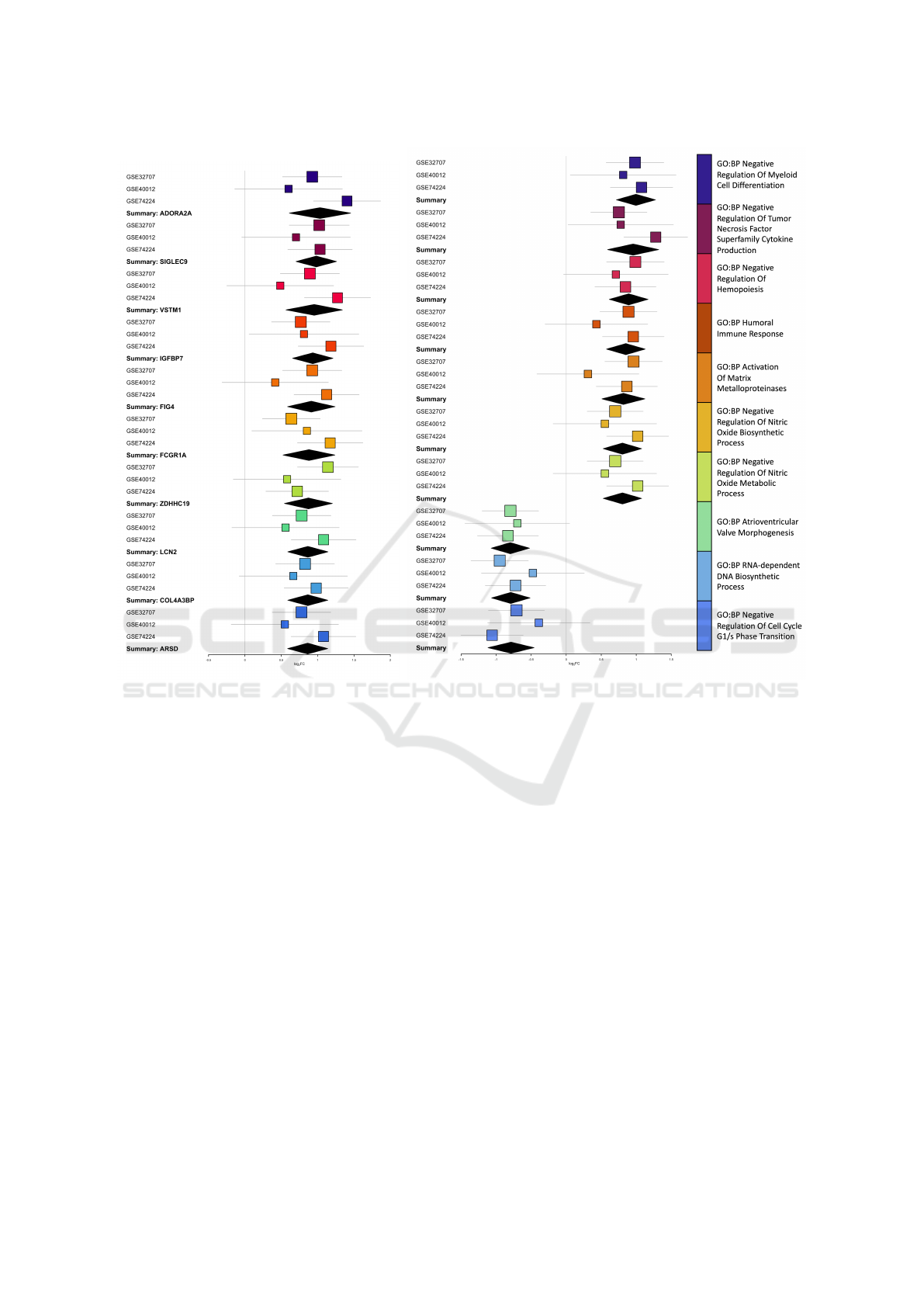
(a) (b)
Figure 1: The top 10 associated genes and pathways from from the filtered meta analysis results.
unique validation datasets does suggest that a path-
way based signature is clinically useful in the devel-
opment of molecular biomarker signatures.
While pathways are more reproducible compared
to genes in this work, much of previous work in
molecular diagnostics has focused on a minimal set
of biomarkers to be measured in order to minimize di-
agnostic costs (Sweeney et al., 2015). However, due
to the falling costs of whole genome RNA sequenc-
ing, we are approaching a time when sequencing the
entire transcriptome will be as cost effective, if not
cheaper, then targeted technologies. Such approaches
open up the possibility of using these pathway based
signatures without additional costs.
One of common critiques of the use of AI in di-
agnosis is a lack of clinical interpretability, with clin-
icians feeling uncomfortable with the ‘black box’ ap-
proach to diagnosis. While pathway signatures cannot
serve to ‘lift the hood’ under complicated AI algo-
rithms, they do allow researchers, clinicians, and pa-
tients to better understand the biological inputs under-
pinning the diagnostic prediction models (Wang et al.,
2020). As the pathways here are generated from the
ranked DEGs, the meta-score pathway signature both
preserves more information that just DEGs while also
increasing interpretability of biomarkers.
Sepsis is characterized by dysregulation in the
host immune system and inflammatory response,
which then causes severe oxidative stress.(Macdonald
et al., 2003) Excessive free radicals or inadequate de-
fenses can cause lipid peroxidation, impact cell and
mitochondrial membrane stability and lead to cell
death and tissue damage. (Macdonald et al., 2003;
Fanucchi, 2014) Accordingly, we would expect to
see pathways involved in immune response, heat and
oxidative stress, and apoptosis. Nuclear factor κB
(NFκB) is a ubiquitous transcription factor that is
thought to regulate innate immunity and be involved
in inflammation, cancer, and nervous system func-
tion. (Macdonald et al., 2003; Salminen et al., 2008;
Albensi, 2019) By over-regulating downstream pro-
teins, this transcription factor may contribute to the
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
164

Figure 2: The gene and pathway meta-score ROC curves in
the validation datasets.
immune dysregulation in sepsis. Several of the path-
ways included in our signature are thought to directly
or indirectly regulate or be regulated by NFκB, in-
cluding: Negative regulation of myeloid cell differen-
tiation (Achyut et al., 2017), Cell killing (Fan et al.,
2008), Negative regulation of tumor necrosis factor
superfamily cytokine production (Hayden and Ghosh,
2014), Regulation of fatty acid metabolism (Kracht
et al., 2020), Polyamine biosynthetic process (Fac-
chini et al., 2005), Regulation of cell cycle G1/S phase
transition (Ledoux and Perkins, 2014). The tight in-
terconnection of these pathways supports the biolog-
ical relevance of our signature, and suggests the rela-
tionship between these processes as a target for future
research in sepsis.
4 CONCLUSION
This work, while promising, is not without limita-
tion. Future work should consider whether the in-
creases seen in reproducibility of biomarkers is true
across many diseases and compare across both mi-
croarray and RNA sequencing. As well, we sought
only to validate these findings in datasets collected
on the same tissue. Additional work should consider
whether pathway signatures are more reproducible
across different tissue types compared to gene ex-
pression signatures. Moreover, comparing the perfor-
mance of pathway based approaches in different diag-
nostic models should be considered.
We demonstrate that pathway signatures are more
replicable than gene signatures and that pathway sig-
natures can be easily amended to existing signature
identification models to improve validation accuracy
in new datasets. Future work emphasizing meth-
ods for pathway-based signatures should occur as
RNA sequencing costs fall and the possibility of cost-
effective pathway signatures becomes reality.
REFERENCES
Aare, S., Radell, P., Eriksson, L. I., Chen, Y.-W., Hoffman,
E. P., and Larsson, L. (2012). Role of sepsis in the
development of limb muscle weakness in a porcine
intensive care unit model. Physiological genomics,
44(18):865–877.
Achyut, B. R., Angara, K., Jain, M., Borin, T. F., Rashid,
M. H., Iskander, A. S. M., Ara, R., Kolhe, R., Howard,
S., Venugopal, N., Rodriguez, P. C., Bradford, J. W.,
and Arbab, A. S. (2017). Canonical nfκb signaling in
myeloid cells is required for the glioblastoma growth.
Scientific Reports, 7(1):13754.
Albensi, B. C. (2019). What is nuclear factor kappa b (nf-
κb) doing in and to the mitochondrion? Frontiers in
Cell and Developmental Biology, 7:154.
Alpern, D., Gardeux, V., Russeil, J., Mangeat, B., Meireles-
Filho, A. C. A., Breysse, R., Hacker, D., and De-
plancke, B. (2019). BRB-seq: ultra-affordable high-
throughput transcriptomics enabled by bulk RNA bar-
coding and sequencing. Genome Biology, 20(1):71.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P.,
Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese,
J. C., Richardson, J. E., Ringwald, M., Rubin, G. M.,
and Sherlock, G. (2000). Gene Ontology: tool for the
unification of biology. Nature Genetics, 25(1):25–29.
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim,
I. F., Tomashevsky, M., Marshall, K. A., Phillippy,
K. H., Sherman, P. M., Holko, M., Yefanov, A., Lee,
H., Zhang, N., Robertson, C. L., Serova, N., Davis,
S., and Soboleva, A. (2012). NCBI GEO: archive for
functional genomics data sets—update. Nucleic Acids
Research, 41(D1):D991–D995.
Bhatty, M., Fan, R., Muir, W. M., Pruett, S. B., and Nan-
duri, B. (2012). Transcriptomic analysis of peritoneal
cells in a mouse model of sepsis: confirmatory and
novel results in early and late sepsis. BMC genomics,
13(1):1–13.
Busse, H., Bitzinger, D., H
¨
ocherl, K., Seyfried, T., Gruber,
M., Graf, B. M., and Zausig, Y. A. (2016). Adeno-
sine a2a and a2b receptor substantially attenuate is-
chemia/reperfusion injury in septic rat hearts. Cardio-
vascular drugs and therapy, 30(6):551–558.
Chen, J. J., Tsai, C.-A., Tzeng, S., and Chen, C.-H. (2007).
Gene selection with multiple ordering criteria. BMC
Bioinformatics, 8(1):74.
Cohen, J., Vincent, J.-L., Adhikari, N. K. J., Machado, F. R.,
Angus, D. C., Calandra, T., Jaton, K., Giulieri, S., De-
laloye, J., Opal, S., Tracey, K., van der Poll, T., and
Replicability of Differentially Expressed Genes Versus Biological Pathways Biomarkers in Diagnosing Sepsis
165

Pelfrene, E. (2015). Sepsis: a roadmap for future re-
search. The Lancet Infectious Diseases, 15(5):581–
614.
Coulthard, M. G., Morgan, M., Woodruff, T. M., Aru-
mugam, T. V., Taylor, S. M., Carpenter, T. C., Lack-
mann, M., and Boyd, A. W. (2012). Eph/ephrin signal-
ing in injury and inflammation. The American journal
of pathology, 181(5):1493–1503.
Crow, M., Lim, N., Ballouz, S., Pavlidis, P., and Gillis, J.
(2019). Predictability of human differential gene ex-
pression. Proceedings of the National Academy of Sci-
ences of the United States of America, 116(13):6491–
6500.
Dolinay, T., Kim, Y. S., Howrylak, J., Hunninghake,
G. M., An, C. H., Fredenburgh, L., Massaro, A. F.,
Rogers, A., Gazourian, L., Nakahira, K., Haspel,
J. A., Landazury, R., Eppanapally, S., Christie, J. D.,
Meyer, N. J., Ware, L. B., Christiani, D. C., Ry-
ter, S. W., Baron, R. M., and Choi, A. M. K.
(2012). Inflammasome-regulated Cytokines Are Crit-
ical Mediators of Acute Lung Injury. American
Journal of Respiratory and Critical Care Medicine,
185(11):1225–1234.
Facchini, A., Borz
´
I, R. M., Marcu, K. B., Stefanelli, C.,
Olivotto, E., Goldring, M. B., Facchini, A., and
Flamigni, F. (2005). Polyamine depletion inhibits
nf-κb binding to dna and interleukin-8 production
in human chondrocytes stimulated by tumor necrosis
factor-α. Journal of Cellular Physiology, 204(3):956–
963.
Fan, Y., Dutta, J., Gupta, N., Fan, G., and G
´
elinas, C.
(2008). Regulation of programmed cell death by nf-
κb and its role in tumorigenesis and therapy. In Pro-
grammed Cell Death in Cancer Progression and Ther-
apy, volume 615, pages 223–250. Springer Nether-
lands, Dordrecht. Series Title: Advances in Experi-
mental Medicine and Biology.
Fanucchi, M. V. (2014). Development of antioxidant and
xenobiotic metabolizing enzyme systems.
Ferrer, R., Martin-Loeches, I., Phillips, G., Osborn, T. M.,
Townsend, S., Dellinger, R. P., Artigas, A., Schorr,
C., and Levy, M. M. (2014). Empiric Antibiotic
Treatment Reduces Mortality in Severe Sepsis and
Septic Shock From the First Hour: Results From
a Guideline-Based Performance Improvement Pro-
gram*. Critical Care Medicine, 42(8):1749–1755.
Guillen-Guio, B., Lorenzo-Salazar, J. M., Ma, S.-F., Hou,
P.-C., Hernandez-Beeftink, T., Corrales, A., Garc
´
ıa-
Laorden, M. I., Jou, J., Espinosa, E., Muriel, A.,
et al. (2020). Sepsis-associated acute respiratory dis-
tress syndrome in individuals of european ancestry: a
genome-wide association study. The Lancet Respira-
tory Medicine, 8(3):258–266.
H
¨
anzelmann, S., Castelo, R., and Guinney, J. (2013).
GSVA: gene set variation analysis for microarray and
RNA-Seq data. BMC Bioinformatics, 14:7.
Hayden, M. S. and Ghosh, S. (2014). Regulation of nf-
κb by tnf family cytokines. Seminars in Immunology,
26(3):253–266.
Haynes, W. A., Vallania, F., Liu, C., Bongen, E., Tomczak,
A., Andres-Terre, M., Lofgren, S., Tam, A., Deis-
seroth, C. A., Li, M. D., Sweeney, T. E., and Khatri,
P. (2016). Empowering multi-cohort gene expression
analysis to increase reproducibility. Pacific Sympo-
sium on Biocomputing.
Ji, S., Pan, Y., Lu, Q., Sun, Z., and Liu, Y. (2014). Screen-
ing of differentially expressed genes between multiple
trauma patients with and without sepsis. Genet Mol
Res, 13(1):1855–64.
Karp, P. D., Billington, R., Caspi, R., Fulcher, C. A., La-
tendresse, M., Kothari, A., Keseler, I. M., Krummen-
acker, M., Midford, P. E., Ong, Q., Ong, W. K., Paley,
S. M., and Subhraveti, P. (2019). The BioCyc collec-
tion of microbial genomes and metabolic pathways.
Briefings in Bioinformatics, 20(4):1085–1093.
Khatri, P., Sirota, M., and Butte, A. J. (2012). Ten Years
of Pathway Analysis: Current Approaches and Out-
standing Challenges. PLoS Computational Biology,
8(2):e1002375.
Kim, K. S., Jekarl, D. W., Yoo, J., Lee, S., Kim, M., and
Kim, Y. (2021). Immune gene expression networks
in sepsis: A network biology approach. PLOS ONE,
16(3):e0247669.
Kracht, M., M
¨
uller-Ladner, U., and Schmitz, M. L. (2020).
Mutual regulation of metabolic processes and proin-
flammatory nf-κb signaling. Journal of Allergy and
Clinical Immunology, 146(4):694–705.
Ledoux, A. and Perkins, N. (2014). Nf-κb and the cell cy-
cle. Biochemical Society Transactions, 42(1):76–81.
Li, Z., Zhang, Y., Liu, Y., Liu, Y., and Li, Y. (2017). Iden-
tification of key genes in gram-positive and gram-
negative sepsis using stochastic perturbation. Molec-
ular medicine reports, 16(3):3133–3146.
Lim, K., Li, Z., Choi, K. P., and Wong, L. (2015). A quan-
tum leap in the reproducibility, precision, and sensi-
tivity of gene expression profile analysis even when
sample size is extremely small. Journal of Bioinfor-
matics and Computational Biology, 13(04):1550018.
Macdonald, J., Galley, H., and Webster, N. (2003). Ox-
idative stress and gene expression in sepsis. British
Journal of Anaesthesia, 90(2):221–232.
Martens, M., Ammar, A., Riutta, A., Waagmeester, A.,
Slenter, D., Hanspers, K., A. Miller, R., Digles, D.,
Lopes, E., Ehrhart, F., Dupuis, L. J., Winckers, L. A.,
Coort, S., Willighagen, E. L., Evelo, C. T., Pico, A. R.,
and Kutmon, M. (2021). WikiPathways: connecting
communities. Nucleic Acids Research, 49(D1):D613–
D621.
Mayday, M. Y., Khan, L. M., Chow, E. D., Zinter, M. S., and
DeRisi, J. L. (2019). Miniaturization and optimiza-
tion of 384-well compatible RNA sequencing library
preparation. PLOS ONE, 14(1):e0206194.
McHugh, L., Seldon, T. A., Brandon, R. A., Kirk, J. T.,
Rapisarda, A., Sutherland, A. J., Presneill, J. J., Ven-
ter, D. J., Lipman, J., Thomas, M. R., Klein Klouwen-
berg, P. M. C., van Vught, L., Scicluna, B., Bon-
ten, M., Cremer, O. L., Schultz, M. J., van der Poll,
T., Yager, T. D., and Brandon, R. B. (2015). A
Molecular Host Response Assay to Discriminate Be-
tween Sepsis and Infection-Negative Systemic Inflam-
mation in Critically Ill Patients: Discovery and Val-
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
166

idation in Independent Cohorts. PLOS Medicine,
12(12):e1001916.
Mootha, V. K., Lindgren, C. M., Eriksson, K.-F., Subrama-
nian, A., Sihag, S., Lehar, J., Puigserver, P., Carls-
son, E., Ridderstr
˚
ale, M., Laurila, E., Houstis, N.,
Daly, M. J., Patterson, N., Mesirov, J. P., Golub,
T. R., Tamayo, P., Spiegelman, B., Lander, E. S.,
Hirschhorn, J. N., Altshuler, D., and Groop, L. C.
(2003). PGC-1α-responsive genes involved in oxida-
tive phosphorylation are coordinately downregulated
in human diabetes. Nature Genetics, 34(3):267–273.
Parnell, G. P., McLean, A. S., Booth, D. R., Armstrong,
N. J., Nalos, M., Huang, S. J., Manak, J., Tang, W.,
Tam, O.-Y., Chan, S., and Tang, B. M. (2012). A
distinct influenza infection signature in the blood tran-
scriptome of patients with severe community-acquired
pneumonia. Critical Care, 16(4):R157.
Pradines, J. R., Farutin, V., Cilfone, N. A., Ghavami, A.,
Kurtagic, E., Guess, J., Manning, A. M., and Capila,
I. (2020). Enhancing reproducibility of gene expres-
sion analysis with known protein functional relation-
ships: The concept of well-associated protein. PLOS
Computational Biology, 16(2):e1007684.
R Core Team (2017). R: A Language and Environment for
Statistical Computing. R Foundation for Statistical
Computing, Vienna, Austria.
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi,
W., and Smyth, G. K. (2015). limma powers differen-
tial expression analyses for RNA-sequencing and mi-
croarray studies. Nucleic Acids Research, 43(7):e47–
e47.
Salminen, A., Huuskonen, J., Ojala, J., Kauppinen, A.,
Kaarniranta, K., and Suuronen, T. (2008). Activation
of innate immunity system during aging: NF-kB sig-
naling is the molecular culprit of inflamm-aging. Age-
ing Research Reviews, 7(2):83–105.
Scicluna, B. P., Klein Klouwenberg, P. M. C., van Vught,
L. A., Wiewel, M. A., Ong, D. S. Y., Zwinderman,
A. H., Franitza, M., Toliat, M. R., N
¨
urnberg, P.,
Hoogendijk, A. J., Horn, J., Cremer, O. L., Schultz,
M. J., Bonten, M. J., and van der Poll, T. (2015).
A Molecular Biomarker to Diagnose Community-
acquired Pneumonia on Intensive Care Unit Admis-
sion. American Journal of Respiratory and Critical
Care Medicine, 192(7):826–835.
Sholder, G., Lanz, T. A., Moccia, R., Quan, J., Aparicio-
Prat, E., Stanton, R., and Xi, H. S. (2020). 3’Pool-seq:
an optimized cost-efficient and scalable method of
whole-transcriptome gene expression profiling. BMC
Genomics, 21(1):64.
Subramanian, A., Tamayo, P., Mootha, V. K., Mukher-
jee, S., Ebert, B. L., Gillette, M. A., Paulovich, A.,
Pomeroy, S. L., Golub, T. R., Lander, E. S., and
Mesirov, J. P. (2005). Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-
wide expression profiles. Proceedings of the National
Academy of Sciences, 102(43):15545–15550.
Sutherland, A., Thomas, M., Brandon, R. A., Brandon,
R. B., Lipman, J., Tang, B., McLean, A., Pascoe,
R., Price, G., Nguyen, T., Stone, G., and Venter, D.
(2011). Development and validation of a novel molec-
ular biomarker diagnostic test for the early detection
of sepsis. Critical Care, 15(3):R149.
Sweeney, T. E. and Khatri, P. (2017). Benchmarking Sep-
sis Gene Expression Diagnostics Using Public Data*:.
Critical Care Medicine, 45(1):1–10.
Sweeney, T. E., Shidham, A., Wong, H. R., and Khatri, P.
(2015). A comprehensive time-course–based multico-
hort analysis of sepsis and sterile inflammation reveals
a robust diagnostic gene set. Science Translational
Medicine, 7(287):287ra71–287ra71.
Tan, P. K. (2003). Evaluation of gene expression measure-
ments from commercial microarray platforms. Nu-
cleic Acids Research, 31(19):5676–5684.
Tweedie, S., Braschi, B., Gray, K., Jones, T. E. M., Seal, R.,
Yates, B., and Bruford, E. A. (2021). Genenames.org:
the HGNC and VGNC resources in 2021. Nucleic
Acids Research, 49(D1):D939–D946.
van Zanten, A. R. H. (2014). The Golden Hour of Antibi-
otic Administration in Severe Sepsis: Avoid a False
Start Striving for Gold*. Critical Care Medicine,
42(8):1931–1932.
von Gunten, S., Jakob, S. M., Geering, B., Takala, J., and
Simon, H.-U. (2009). Different patterns of siglec-9-
mediated neutrophil death responses in septic shock.
Shock, 32(4):386–392.
Wang, F., Kaushal, R., and Khullar, D. (2020). Should
health care demand interpretable artificial intelligence
or accept “black box” medicine?
Wasyluk, W. and Zwolak, A. (2021). Parp inhibitors: An
innovative approach to the treatment of inflammation
and metabolic disorders in sepsis. Journal of Inflam-
mation Research, 14:1827.
Webber, W., Moffat, A., and Zobel, J. (2010). A similarity
measure for indefinite rankings. ACM Transactions on
Information Systems, 28(4):1–38.
Wu, G. and Haw, R. (2017). Functional Interaction Net-
work Construction and Analysis for Disease Discov-
ery. In Wu, C. H., Arighi, C. N., and Ross, K. E.,
editors, Protein Bioinformatics, volume 1558, pages
235–253. Springer New York, New York, NY. Series
Title: Methods in Molecular Biology.
Zarringhalam, K., Enayetallah, A., Reddy, P., and Ziemek,
D. (2014). Robust clinical outcome prediction based
on Bayesian analysis of transcriptional profiles and
prior causal networks. Bioinformatics, 30(12):i69–
i77.
Zhang, M., Yao, C., Guo, Z., Zou, J., Zhang, L., Xiao, H.,
Wang, D., Yang, D., Gong, X., Zhu, J., Li, Y., and Li,
X. (2008). Apparently low reproducibility of true dif-
ferential expression discoveries in microarray studies.
Bioinformatics, 24(18):2057–2063.
Zhang, M., Zhang, L., Zou, J., Yao, C., Xiao, H., Liu,
Q., Wang, J., Wang, D., Wang, C., and Guo, Z.
(2009). Evaluating reproducibility of differential ex-
pression discoveries in microarray studies by consid-
ering correlated molecular changes. Bioinformatics,
25(13):1662–1668.
Replicability of Differentially Expressed Genes Versus Biological Pathways Biomarkers in Diagnosing Sepsis
167
