
Skin Cancer Classification using Deep Learning Models
Marwa Kahia
a
, Amira Echtioui
b
, Fathi Kallel
c
and Ahmed Ben Hamida
d
ATMS Lab, Advanced Technologies for Medicine and Signals, ENIS, Sfax University, Sfax, Tunisia
Keywords: Melanoma, Diagnosis, VGG16, Skin Cancer, InceptionV3.
Abstract: In recent years, researches proved that Melanoma is the deadliest form of skin cancer. In the early stages, it
can be treated successfully with surgery alone and survival rates are high. A large number of methods for
Melanoma classification has been proposed to deal with this problem, but although they did not find better
ways to create the final solution. Thus, our aim is to go further and explore the classic models in order to
handle the Melanoma classification problem based on modified VGG16 and modified InceptionV3. The
conducted experiments revealed the effectiveness of our proposed method based on modified VGG16 with
73.33% of accuracy, when compared to other state-of-the-art methods on the same data sets, in terms of
finding optimal and effective solutions and improving the objective function.
1 INTRODUCTION
Melanoma is the most unsafe form of skin cancer. It
begins in the melanocytes (color- producing cells
plant in the surface subcaste of the skin). In the utmost
of cases, it's caused by ultraviolet radiation from sun
or tanning beds which produce mutations (inheritable
blights) that take the skin cells to expand fleetly and
form nasty excrescences (l. Argenziano, et al., 2000).
Melanoma causes 55 500 cancer deaths annually
which is 0.7 of all cancer deaths. The prevalence and
mortality rates of carcinoma differ from one country
to another due to the variation of ethnical and ethnical
groups (Schadendorf et al., 2018). Nasty carcinoma is
presumptive to come one of the most common nasty
excrescences in the future, with yet a ten times
advanced prevalence rate (Tadeusiewicz et al., 2010).
Visual examination of the suspicious skin area is
generally adopted by dermatologist as a first step for
the diagnosis of a malignant lesion. In fact, an
accurate diagnosis is essential because of the
resemblances of some lesion types. Furthermore, the
diagnostic accuracy correlates strongly with the
professional experience of the physician
(Tadeusiewicz et al., 2010).
On the other hand, without any further technical
support, dermatologists have a 65% to 80% accuracy
a
https://orcid.org/0000-0001-5255-9568
b
https://orcid.org/0000-0003-2041-1301
c
https://orcid.org/0000-0001-7986-8395
d
https://orcid.org/0000-0001-6713-7384
rate in melanoma diagnosis. In suspicious cases,
dermatologists explore and use dermatoscopic
images as a complementary support of the visual
inspection. In fact, the combination of both visual
inspection and dermatoscopic images eventually
results in an absolute melanoma detection accuracy of
75%-84% by dermatologists (Brinker et al., 2018)
Currently, artificial intelligence (AI) has come an
aptitude to face these problems. Several deep-literacy
infrastructures like reccurent neural networks (RNN),
convolutional neural networks (CNN), deep neural
networks (DNN), long short term memory (LSTM) are
proposed in literature to descry cancer cell. These
models are also successfully performed in classifying
skin cancer.
Several CNN architectures, like ResNet,
Inception and Xception, as well as VGG16, are
proposed in literature and specially designed for
image classification. Numerous researchers have
developed methods based on deep learning to classify
and identify skin cancer (Le et al., 2020; Garg et al.,
2019; Guan et al., 2019; Nugroho et al., 2019;
Pacheco et al., 2019).
In this work, we propose a modified InceptionV3
model for the classification of skin cancer. We
propose also a modified VGG16 model which
classifies skin cancer with a better accuracy value
554
Kahia, M., Echtioui, A., Kallel, F. and Ben Hamida, A.
Skin Cancer Classification using Deep Learning Models.
DOI: 10.5220/0010976400003116
In Proceedings of the 14th International Conference on Agents and Artificial Intelligence (ICAART 2022) - Volume 1, pages 554-559
ISBN: 978-989-758-547-0; ISSN: 2184-433X
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
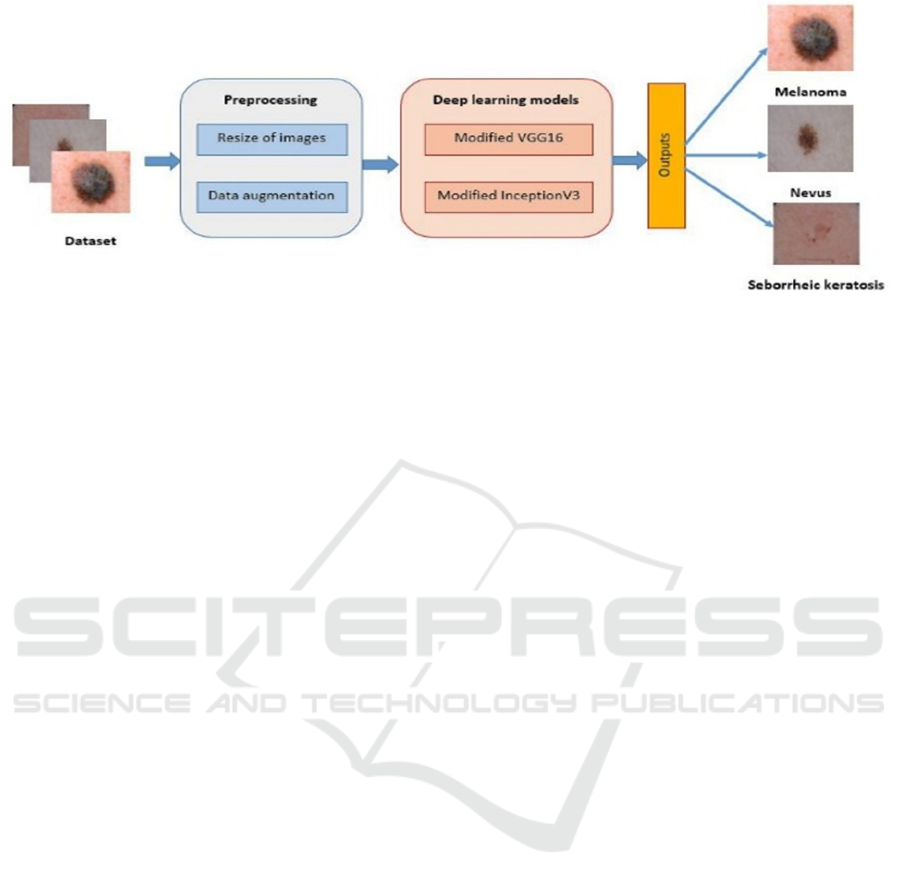
Figure 1: Flowchart of the proposed method for skin cancer classification using modified VGG16 model.
compared to the state of the art.
The rest of the paper is organised as follows:
Section 2 details materials and proposed method.
Section 3 represents results and discussion. Section 4
concludes this paper.
2 MATERIAL AND PROPOSED
METHOD
In this section, we will present the dataset used in this
research work and present our proposed method for
skin cancer classification.
2.1 Dataset Description
The used dataset in this present work contains three
classes: melanoma, nevus and seborrheic keratosis.
More details about this datasets are given below:
2000 training images
(https://s3-us-west-1.amazonaws.com/udacity-
dlnfd/datasets/skin-cancer/train.zip)
- melanoma images: 374
- nevus images: 1372
- seborrheic keratosis images: 254
150 validation images
(https://s3-us-west-1.amazonaws.com/udacity-
dlnfd/datasets/skin-cancer/valid.zip)
600 testing images
(https://s3-us-west-1.amazonaws.com/udacity-
dlnfd/datasets/skin-cancer/test.zip)
2.2 Proposed Method
Figure 1 presents Flowchart of the proposed method.
A preprocessing stage is firstly applied on input
image. The preprocessing involves resizing all
images and increasing the number of images from
both classes melanoma and seborrheic keratosis.
Then we test the modified VGG16 model and apply
our modified InceptionV3 model.
2.2.1 Data Augmentation
We used data augmentation techniques to artificially
boost the amount of our training data because our
data collection is rather small. The increase in data is
an often-applied DL method that generates the
required number of samples. It also improves
network efficiency for a small database by
optimizing it. Shifting, Rotation, flipping,
transformation, and zooming are all examples of
traditional data augmentation procedures. We used
“Keras Image Data Generator” to apply image
augmentations during training in this investigation.
As shown in section 2.1, the number of images of
class 'Nevus' is 1372. In order to balance the number
of images for all three considered classes, we applied
the data augmentation technique to augment the size
of both classes 'Melanoma' and 'seborrheic keratosis'.
In this work, we choose a vertical flip, a
horizontal flip and a 45-degree rotation for data
augmentation. As a result, we got 1372 images for
each class.
2.2.2 Skin Cancer Classification using
Modified VGG16 Model
Figure 2 shows the flowchart of the proposed method
for the classification of skin cancer using the VGG16
model. In this paper, modified VGG16 begin by five
blocks, the first two blocks include two convolutional
layers with a Relu activation function and Max
Pooling followed by three blocks. Each block enclose
three convolutional layers with a Relu activation
function and Max Pooling. An adaptative Avg
Skin Cancer Classification using Deep Learning Models
555
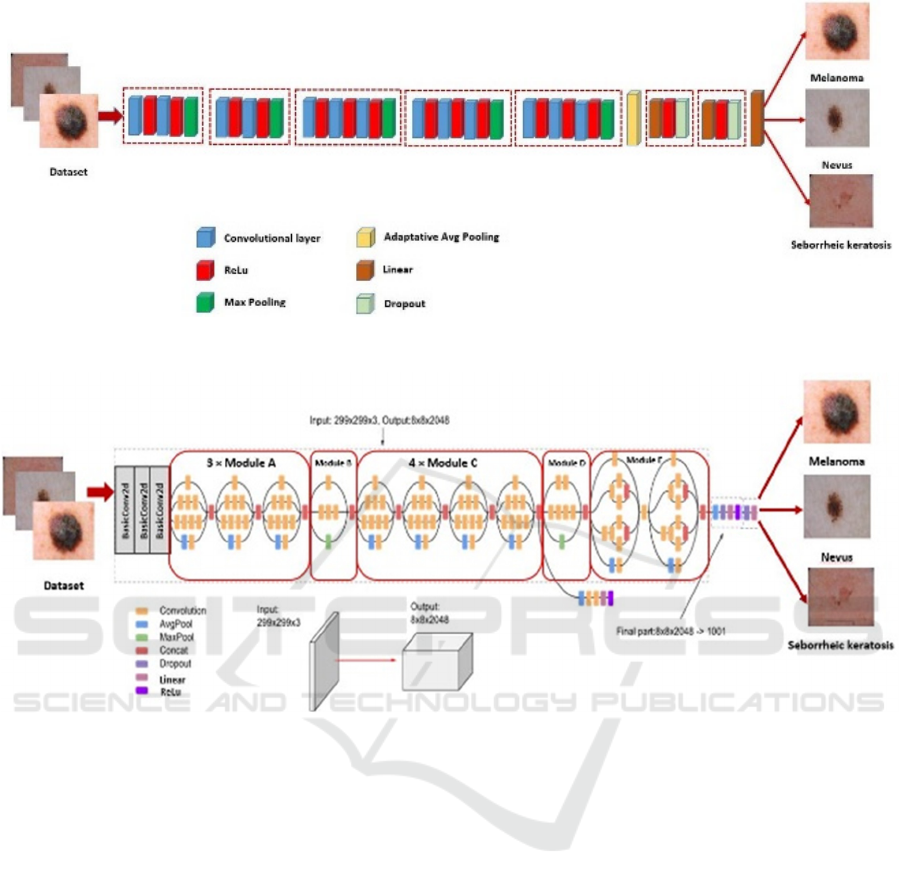
Figure 2: Flowchart of the modified VGG16 for skin cancer classification.
Figure 3: Flowchart of the modified InceptionV3 for skin cancer classification.
Pooling and two blocks follow these blocks. Each
block contains linear layer, ReLu activation function,
and Dropout Layer. Finally, a linear layer is used to
predict the class of images.
We fine-tuned this model by 10 epochs. The
Adaptive Moment Estimation known as “Adam
optimizer” is used to optimize the loss function. The
adopted model is trained by a cross-entropy loss
function.
2.2.3 Skin Cancer Classification using
Modified InceptionV3 Model
Figure 3 shows the modified method for the
classification of skin cancer using the InceptionV3
model. InceptionV3 is a commonly used image
classification model that has demonstrated more than
78.1% accuracy on the ImageNet dataset. The model
itself is made up of basic symmetric and asymmetric
components including convolutions, average pooling,
maximum pooling, concatenations, drops, and fully
connected layers. Batch normalization is widely used
in the model and applied to activation inputs. The loss
is calculated via SoftMax. Our Modified InceptionV3
begins by three blocks of BasicConv2d. Each block
includes a convolutional layer and a batch
normalization step followed by 3 Modules A, module
B, 4 modules C, module D, and 2 modules E followed
by Avg Pooling, Dropout, Linear layer, ReLu,
Dropout layer and Linear layer.
3 RESULTS AND DISCUSSION
In this section, we present and discuss the obtained
classification results when both proposed models are
used. Accuracy, precision, recall and F1-score
metrics are considered for performance evaluation of
proposed classifiers. These mentioned metrics are
respectively computed according to the following
SDMIS 2022 - Special Session on Super Distributed and Multi-agent Intelligent Systems
556
equations for both modified VGG16 and modified
InceptionV3 models.
𝑎𝑐𝑐𝑢𝑟𝑎𝑐𝑦
(1)
𝑝𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛
(2)
𝑟𝑒𝑐𝑎𝑙𝑙
(3)
𝐹1 𝑠𝑐𝑜𝑟𝑒 2 ∗
∗
(4)
where TP, TN, FP and FN are respectively the True
Positive, True Negative, False Positive and False
Negative.
Both modified VGG16 and modified InceptionV3
algorithms assess the classification performance. We
achieved two experiments using the same described
dataset. We conducted the first classification
experiment considering all melanoma, nevus and
Seborrheic keratosis classes. The second
classification experiment is executed considering
only two classes: benign and malignant classes.
3.1 Classification Results: Three
Classes
In this section, we present the obtained classification
results when the three classes are considered. Table 1
presents the average accuracy results of all considered
classes for both modified VGG16 and modified
InceptionV3 models.
Table 1: Classification accuracy.
Accuracy
Modified VGG16
73.33%
Modified InceptionV3
42.00%
Table 2 details the accuracy results obtained with
three considered classes for both modified VGG16
and modified InceptionV3 models.
Table 2: Classification accuracy for three classes.
Modified
VGG16
Modified
InceptionV3
melanoma 50% 33%
nevus 54% 84%
Seborrheic keratosis 47% 24%
From tables 1, we can observe that modified
VGG16 model performs better than the modified
InceptionV3 model. In fact, the average accuracy
value obtained with modified VGG16 model is better
(73.33%) than those obtained with modified
InceptionV3 model (only 42%).
Table 2 showed that both proposed methods
present good classification performances for 'Nevus'
class with a superiority for modified InceptionV3
model. In fact, this class achieves an accuracy value
of 54% with modified VGG16 and 84% with modified
InceptionV3. However, classification performances
using both proposed methods are significantly
decreased for 'Seborrheic keratosis' class. In this
case, accuracy values are only limited to 47% and
24% for modified VGG16 and modified InceptionV3
models respectively.
3.2 Classification Results: Two Classes
In this section, we present the obtained classification
results when the two benign and malignant classes are
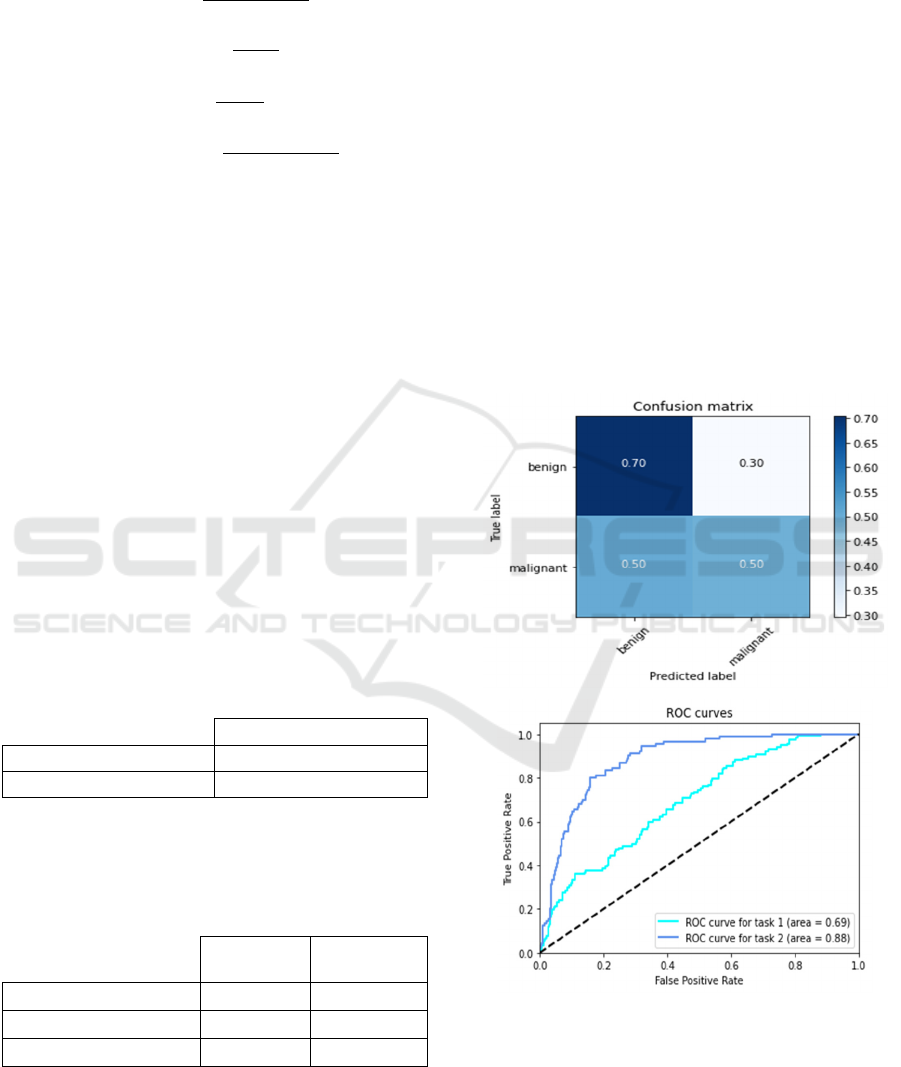
considered. Figure 4 shows the confusion matrix and
the ROC curves for both Modified VGG16 model.
Figure 4: Confusion matrix and ROC curve for modified
VGG16 model.
Figure 5 shows the confusion matrix and the ROC
curves for both Modified InceptionV3 model.
Skin Cancer Classification using Deep Learning Models
557
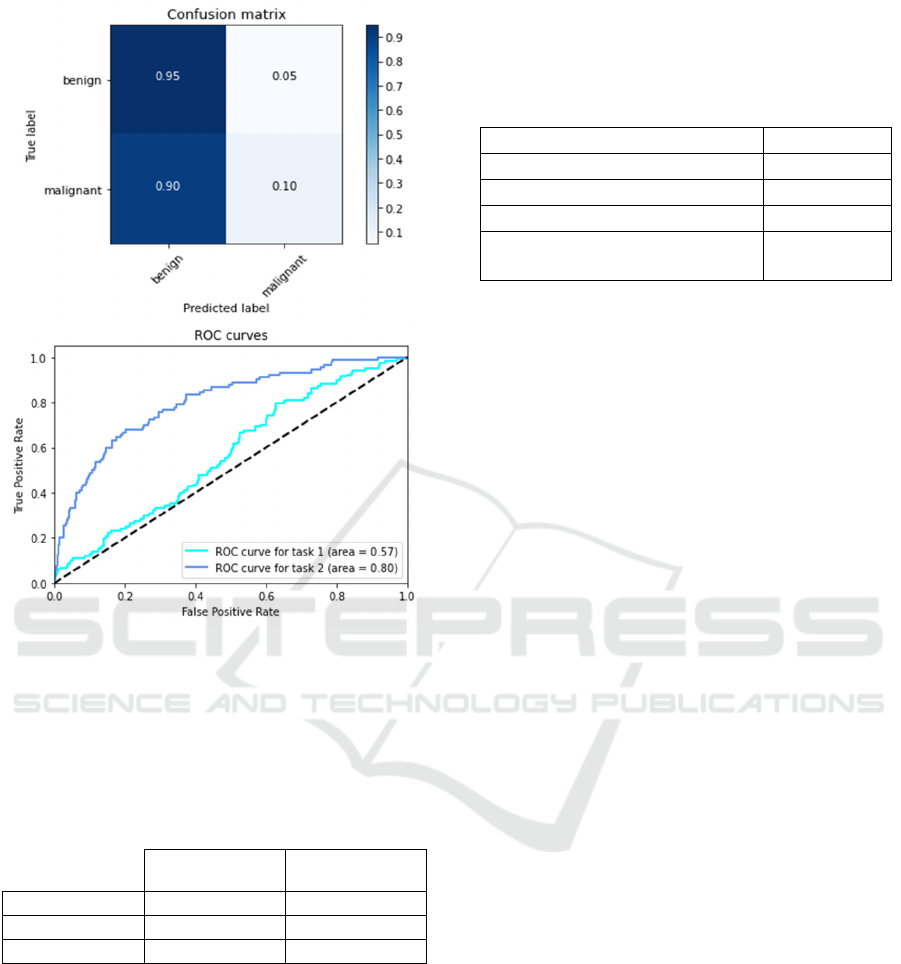
Figure 5: Confusion matrix and ROC curve for modified
InceptionV3 model.
Table 3 reports the average results for recall,
precision and F1-score metrics computed using both
proposed VGG16 and InceptionV3 models.
Table 3: Classification performances for Malignant and
Benign classes.
Modified
VGG16
Modified
Ince
p
tionV3
Recall 51.35% 58.33%
Precision 95.00% 70.00%
F1-score 66.66% 63.63%
The binary classification of Malignant and Benign
classes also show that the proposed method based on
the VGG16 model achieves better performances then
the second proposed method based on InceptionV3
model. In fact, considering the proposed VGG16
model, recall, precision and F1-score values are
respectively equal to 51.35%, 95.00%, and 66.66%.
3.3 Discussion
The performances of the modified VGG16 model are
compared to three state of the art methods labelled as
KNN (Daghrir et al., 2020), SVM (Daghrir et al.,
2020) and AlexNet (Sasikala et al., 2020). Results are
summarized in Table 4.
Table 4: Comparative study for binary classification.
Method Accuracy
KNN (Da
g
hrir et al., 2020) 57.3%
SVM (Daghrir et al., 2020) 71.8%
AlexNet (Sasikala et al., 2020) 65.3 %
Proposed method based on
modified VGG16
73.33%
By comparing the accuracy values listed in Table
4 obtained for different considered methods, we can
observe that our modified VGG16 method performs
better than KNN, SVM, and AlexNet methods. In fact,
accuracy reached 73.33% with our proposed VGG16
method. Although the accuracy is limited to 57.3%,
the KNN method is able to hardly identify malignant
skin lesions since it is sensitive to outliers.
On the other hand, the SVM method performs
better than the KNN and AlexNet methods due to its
adaptability and efficiency. In fact, accuracy is equal
to 71.8% with SVM method, but it is limited to only
57.3% and 65.3% with KNN and AlexNet methods
respectively. Although AlexNet achieved quiet
performance, the SVM is still considered a more
robust and powerful tool for identifying skin cancer.
4 CONCLUSIONS
In this work, we proposed two modified models for
skin cancer classification: modified VGG16 and
modified InceptionV3 models. The application of the
data augmentation showed that the reduction of the
data imbalance can be useful to improve classification
performance, but careful tuning is required, for
example, to make the data perfectly balanced training
does not necessarily result in a better model.
Performances are evaluated using different
metrics like accuracy, precision, recall and F1-score.
Two experiments are conducted. In the first
experiment, we considered melanoma, nevus and
Seborrheic keratosis classes, but in the second one,
only benign and malignant classes are considered.
Results of first experiment showed that the modified
VGG16 is a reliable multiple classifier and performs
better than modified InceptionV3 model. For second
experiment, compared to state of the art considered
methods, results showed that better accuracy values
are obtained for binary classification using modified
VGG16 model.
SDMIS 2022 - Special Session on Super Distributed and Multi-agent Intelligent Systems
558

It is clear that our proposed method given better
results compared to different others recent methods.
However, there is a need to improve its performances
in our future work. In fact, merging or concatenating
deep learning models could improve the classification
results.
REFERENCES
Brinker, T.S., Hekler, A., Utikal, J., Grabe, N.,
Schadendorf, D., Klode, J., Berking, C., Steeb, T., Enk,
A., & von Kalle, C., Skin Cancer Classification Using
Convolutional Neural Networks: Systematic Review J
Med Internet Res. 2018 Oct; 20(10): e11936
Daghrir, J., Tlig, L., Bouchouicha, M., Sayadi, M.
Melanoma skin cancer detection using deep learning
and classical machine learning techniques: A hybrid
approach. International Conference on Advanced
Technologies for Signal and Image Processing, Sep
2020.
Garg, R., Maheshwari, S., & Shukla, A. (2019). Decision
support system for detection and classification of skin
cancer using CNN. arXiv preprint arXiv:1912.03798.
Guan, Q., Wang, Y., Ping, B., Li, D., Du, J., Qin, Y., et al.
(2019). Deep convolutional neural network VGG-16
model for differential diagnosing of papillary thyroid
carcinomas in cytological images: a pilot study. Journal
of Cancer, 10(20), 4876.
Argenziano, G., Soyer H. P., De Giorgio, V., et al.
Interactive Atlas of Dermoscopy Milano, Italy: Edra
Medical Publishing and New Media; 2000.
Le, D. N., Le, H. X., Ngo, L. T., & Ngo, H. T. (2020).
Transfer learning with classweighted and focal loss
function for automatic skin cancer classification. arXiv
preprint arXiv:2009.05977.
Nugroho, A. A., Slamet, I., & Sugiyanto (2019). Skins
cancer identification system of HAMl0000 skin cancer
dataset using convolutional neural network. AIP
Conference Proceedings, 2202, Article 020039.
Pacheco, A. G., & Krohling, R. A. (2019). Recent advances
in deep learning applied to skin cancer detection. arXiv
preprint arXiv:1912.03280.
Sasikala, S., Arun Kumar, S., Shivappriya, S.N. and
Priyadharshini, T. (2020). Towards Improving Skin
Cancer Detection Using Transfer Learning. Biosc.
Biotech. Res. Comm. Special Issue Vol 13 No 11 (2020)
Pp-55-60.
Schadendorf, D., Alexander CJ van Akkooi, Carola
Berking, Klaus G Griewank, Ralf Gutzmer, Axel
Hauschild, Andreas Stang, Alexander Roesch, and
Selma Ugurel. Melanoma. The Lancet,
392(10151):971– 984, 2018.
Tadeusiewicz, R. (2010). Place and role of intelligent
systems in computer science. Computer Methods in
Materials Science;10(4), pp.193-206,2010.
Skin Cancer Classification using Deep Learning Models
559
