
Local Explanations for Clinical Search Engine Results
Edeline Contempr
´
e
a
, Zolt
´
an Szl
´
avik
b
, Majid Mohammadi
c
, Erick Velazquez
d
,
Annette ten Teije
e
and Ilaria Tiddi
f
Vrije Universiteit Amsterdam, De Boelelaan 1105, 1081 HV Amsterdam, The Netherlands
Keywords:
Explainability, Search, Health Care, Crowdsourcing, Treatment Search.
Abstract:
Health care professionals rely on treatment search engines to efficiently find adequate clinical trials and early
access programs for their patients. However, doctors lose trust in the system if its underlying processes are
unclear and unexplained. In this paper, a model-agnostic explainable method is developed to provide users
with further information regarding the reasons why a clinical trial is retrieved in response to a query. To
accomplish this, the engine generates features from clinical trials using by using a knowledge graph, clinical
trial data and additional medical resources. Moreover, a crowd-sourcing methodology is used to determine
features’ importance. Grounded on the proposed methodology, the rationale behind retrieving the clinical trials
is explained in layman’s terms so that healthcare processionals can effortlessly perceive them. In addition, we
compute an explainability score for each of the retrieved items, according to which the items can be ranked.
The experiments validated by medical professionals suggest that the proposed methodology induces trust in
targeted as well as in non-targeted users, and provide them with reliable explanations and ranking of retrieved
items.
1 INTRODUCTION
Health care professionals (HCPs) increasingly rely on
Artificial Intelligence (AI) models to diagnose and,
ultimately, save patients’ lives. In order to use a treat-
ment search engine, HCPs need to gain trust in the
system to find treatment options for their patients.
While accuracy, performance, and design are essen-
tial to ensure trust, more may be needed to reach a
threshold where HCPs trust the system enough to use
it in critical scenarios.
Search engines typically provide a ranked list of
the related items with regards to a certain query. How-
ever, lack of explanations could lead to a lack of trust
from users as they would not understand the underly-
ing logic of retrieving an item in response to a query.
In the medical domain, where the pressure to make no
mistakes is high, incorrectly attributing the cause of a
mistake could be fatal. As a result, without the ability
to interpret the model, HCPs’ trust in the model de-
a
https://orcid.org/0000-0002-1767-121X
b
https://orcid.org/0000-0002-2781-3795
c
https://orcid.org/0000-0002-7131-8724
d
https://orcid.org/0000-0001-9449-8265
e
https://orcid.org/0000-0002-9771-8822
f
https://orcid.org/0000-0001-7116-9338
creases and will, ultimately, not use the model’s out-
puts (Pu and Chen, 2006). In addition, due to com-
pliance regulations, most search engines in the med-
ical domain provide unordered lists of related items
in response to a query, making it difficult for users
to look into or distinguish the most relevant items for
their needs. In addition, for an efficient and thorough
search, a status/date/title-based ordering may not al-
ways be the most practical for end users as they tend
to scatter similar results from each other.
A potential solution to provide explanations to
end-users is to apply current explainable methods.
However, providing HCPs with user-friendly, reli-
able, and easy-to-understand explanations is a cur-
rent challenge in the field. A major drawback of
current explainability techniques, such as LIME (Das
and Rad, 2020), is that these techniques tend to focus
on aiding users with technical backgrounds to inter-
pret the system, and are designed for machine learn-
ing problems such as classification and regression.
HCPs are not universally expected to understand the
detailed workings of a complex retrieval model. Thus,
HCPs require explanations that are high level and un-
derstandable. This opens up an opportunity in which
explanation models can be built in a way that they
are not sensitive to minor changes in the model of a
ContemprÃl’, E., Szlà ˛avik, Z., Mohammadi, M., Velazquez, E., Teije, A. and Tiddi, I.
Local Explanations for Clinical Search Engine Results.
DOI: 10.5220/0010982000003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 735-742
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
735

search engine. For instance, with a treatment search
engine, we may offer a local explanation such as “the
queried disease is mentioned in the retrieved clinical
trial’s title”. Hence, we use a generic explanation
method that can be tailored to individual search en-
gines’ features.
In this paper, an explainability method is devel-
oped which provides tailored explanations to med-
ical practitioners for retrieved items. To that end,
meaningful features from clinical trials are extracted
from different data sources, and preferences of dif-
ferent users are elicited by utilizing a crowdsourcing-
based methodology. We then put forward a method
to translate preferences into importance level of fea-
tures. Based on features’ importance levels, tailored
explanations are acquired for each specific query, ac-
cording to which we develop a sentence template in
order to present them to users. In addition, we in-
troduce explainability scores, according to which we
order retrieved items. The results suggest that the use
of local explainability on clinical search engines pro-
mote HCPs trust, search experience, and result order-
ing satisfaction.
2 RELATED WORK
To clarify our vision of explainability, we identi-
fied three main dimensions of explainability that can
be observed throughout researchers’ definitions: au-
dience, understanding, and transparency. Under-
standing refers to the user’s ability to understand the
model’s results. However, not all users can interpret
all models using explainability as models can be do-
main specific. For example, users without knowledge
in biology would struggle to understand highly bio-
logical terms generated by a model’s explainability
attempting to diagnose a certain type of lung can-
cer. Likewise, explainable AI (XAI) could use simple
terms, leading to a lack of details for the doctor as-
sessing the diagnosis. The user is therefore required
to have a certain amount of knowledge to understand
the explanation itself, making it crucial for developers
using explainability to target their audience (Rosen-
feld and Richardson, 2019). Lastly, an explainable
method should increase the model’s transparency by
making it more interpretable for its users, and not try
to generate seemingly arbitrary explanations that do
not fit with how the model works (Dimanov et al.,
2020).
Current state-of-the-art local explainability tech-
niques do not use user-friendly explanations. These
current local explainability techniques are either
based on feature importance such as LIME (Das and
Rad, 2020) and SHAP (Lundberg and Lee, 2017),
rule-based (Verma and Ganguly, 2019), saliency maps
(Mundhenk et al., 2019), prototypes (Gee et al., 2019)
example based (Dave et al., 2020), or on counterfac-
tual explanations (Dave et al., 2020). Up to date, fea-
ture importance (Zhang et al., 2019) and rule-based
techniques (Verma and Ganguly, 2019) were used on
search engines, but do not meet the criteria that these
should be user friendly.
LIME is a type of local explainability method
aiming to increase transparency for specific decisions
given by an opaque model. It explains single result
by letting users know why they are getting this spe-
cific result over another (Verma and Ganguly, 2019).
Although LIME offers one way to solve the black-
box problem, it has a few limitations. The first lim-
itation of using LIME is that it is most commonly
used for linear or classification models (Arrieta et al.,
2020). This limits the degree to which the model
can be meaningfully applied to, and restricts itself
to non-user-friendly explanations. Consequently, this
research does not use LIME methods, but developed a
local explainability method to order and generate user
friendly explanations.
3 EXPLAINABLE SEARCH
ENGINE
This section presents the proposed model that pro-
vides explanations for its users, as well as how it
orders a clinical search engine’s results. This en-
ables users to efficiently find potential relevant clin-
ical trials while understanding the underlying pro-
cesses of the model. The proposed method also gen-
erates local explainability scores for each clinical trial
and uses these scores to order the search engine’s re-
sults. Moreover, users are provided with user-friendly
explanations delivering descriptions of the features
available in each clinical trial.
Figure 1 shows the pipeline of steps conducted
for the proposed methodology. The search engine
takes as input the user’s query, and returns an out-
put with explainability-based ordered results with ex-
planations. Figure 1 shows that the local explainabil-
ity search engine combines resources with the HCP’s
query to engineer features. These features are, there-
upon, attributed local explainability scores which are
used to order the list of clinical trials. In addition, the
engineered features’ outputs fill template sentences.
These explanations provide information to the user on
how much of this clinical trial can the search engine
explain. In the following sub-sections, each module
in Figure 1 is discussed and explained in more detail.
HEALTHINF 2022 - 15th International Conference on Health Informatics
736
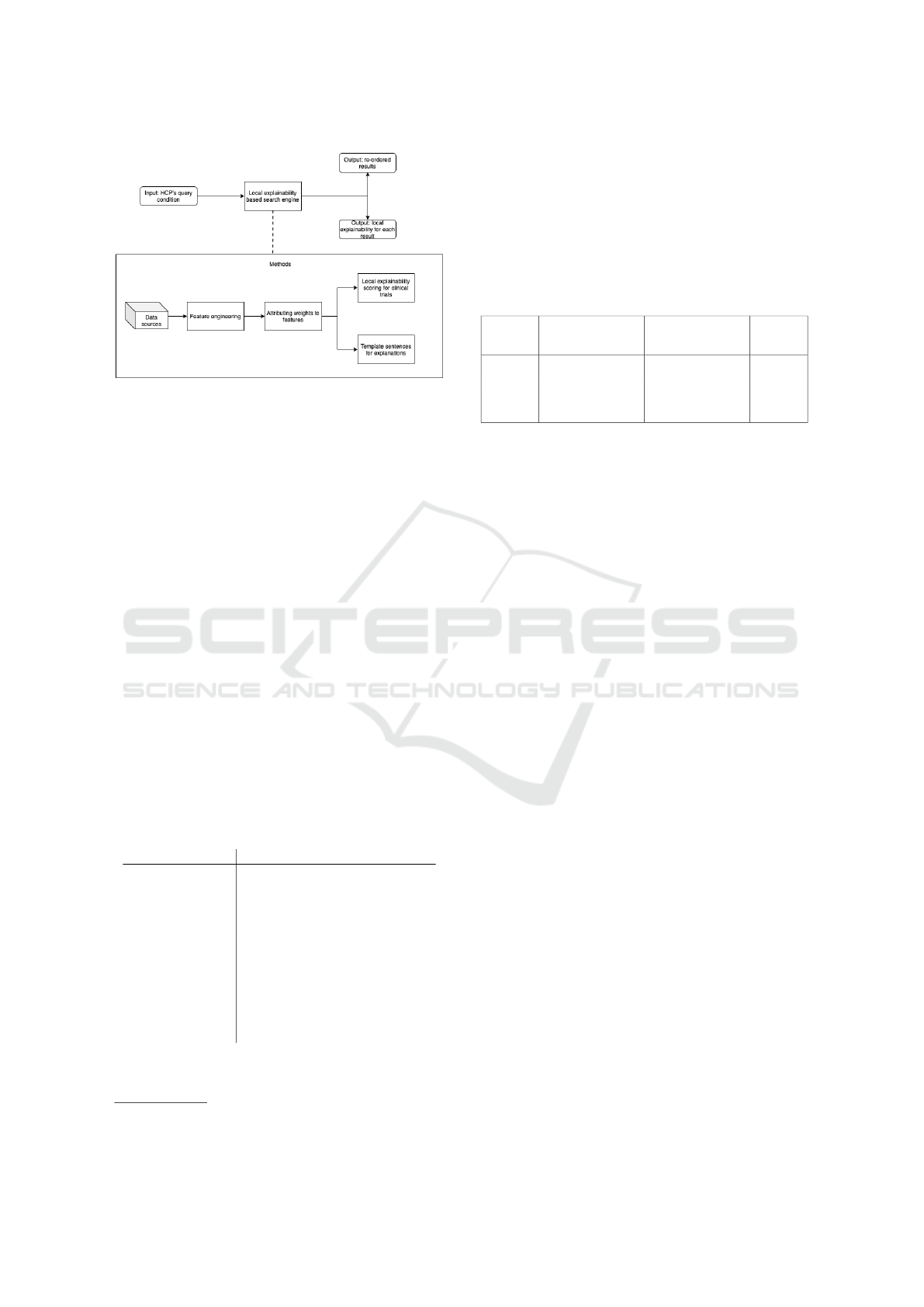
Figure 1: Overview of the methods’ pipeline. Resources in-
clude the knowledge graph, data from UMLS, clinical trials
from CT.gov, and data from Pubmed.
3.1 Feature Engineering
Before engineering the features linked to clinical tri-
als, we first present the data sources according to
which features for each clinical trial are extracted.
Table 1 provides an overview of the data sources
used in the proposed model. First, we used data from
UMLS (Unified Medical Language System) (Boden-
reider, 2004), which is an official medical database
where all conditions, diseases, infections, and more,
are associated to Concept Unique Identifiers (CUIs).
Second, different clinical trial sources such as clini-
caltrials.gov
1
are used as it is the biggest clinical trial
repository. Third, the database comprises Pubmed
publications, as it saves medical papers. Lastly, it
comprises data from the company’s conditions graph
(knowledge graph) where parent-child relations be-
tween diseases are defined, terms are specified, as
well as the clarification of terms and their synonyms.
Table 1: Description of the data sources for the feature en-
gineering.
Data source Data
UMLS Concept Unique Identifiers, disease
terms, and relations between these
clinicaltrials.gov Clinical trials’ detailed descrip-
tions, summaries, phase, title, over-
all status, primary purpose
Pubmed Publications associated to clinical
trials
Knowledge graph Parent-child relationships between
diseases taken from UMLS, dis-
ease concepts (with the diseases’
preferred term and synonyms), lan-
guage.
Table 1 shows the properties, from different data
sources, that were used to engineer features. The
1
https://www.clinicaltrials.gov/
properties, by themselves, do not measure how ex-
plainable a clinical trial is. Therefore, features were
created using the conditions graph, the UMLS and
pubmed databases to assess how much of a clinical
trial the AI can explain. The engineered features are
provided in Table 2.
Table 2: Classification of features created for the local ex-
plainability based search engine.
Feature out-
put type/
Query de-
pendency
Query dependent Query independent Output
Binary query in title, preferred term
in title
clinical stage present, stage
is recruiting, overall status
given
0 or 1
Numeric query in summary, preferred
term in summary, preferred
term in detailed description,
query in detailed description
number of publications Between 0
and infinity
To facilitate the explainability-based calculations,
features were assigned to various categories. Table
2 shows different classifications where the first cate-
gory is based on the user’s query: the feature is either
query dependent or query independent. For example,
the feature query in title is a query dependent feature
as the feature depends on a match between a query
and the title of the clinical trial. In contrast, query
independent features do not depend on the query as
regardless of what the query is, its score remains un-
changed. For example, the feature number of publi-
cations attributed to a clinical trial remains fixed, re-
gardless of user’s query. Second, Table 2 shows en-
gineered features have two distinct outputs which are
either binary or numeric. Binary features assess the
presence of a feature in a study. On the other hand,
numeric features count the occurrence of a feature.
3.2 Feature Importance Identification:
A Crowdsourcing Approach
We created a statistical approach to determine the
weights of our features, and conducted a crowdsourc-
ing task on Amazon Sagemaker as an alternative
method to collect data on feature importance. Com-
pliance regulations prohibits pharmaceutical compa-
nies to retain data on its users, especially when these
relate to drugs. However, previous research has
shown that crowdsourcees provided equal quality an-
swers when conducting medical labeling tasks com-
pared to domain experts (Dumitrache et al., 2013; Du-
mitrache et al., 2017). Hence, in this experiment,
1116 responses were collected from participants to
determine users’ feature preferences and, thereupon,
use these to order and explain results returned by the
clinical search engine.
Features’ importance were measured using a cold
start implicit strategy, where we asked participants to
Local Explanations for Clinical Search Engine Results
737
rate explainability sentences. The rating consisted of
assessing sentences on a 5-point Likert-scale, from
”Not convincing at all” to ”Very convincing”. Each
sentence explained the prominence or availability of
a feature mentioned in Table 2.
To identify user-friendly sentence formulations,
we changed the format of the sentences to implic-
itly measure which sentence format was most pre-
ferred to users. The three formulation dimensions
measured were: numeric vs. non-numeric (using en-
tities ‘3 times’ vs. ‘multiple times’ in an explanatory
sentences), action-oriented versus fact-driven formu-
lations (‘retrieved’ versus ‘clearly mentioned’), and
disease specific versus non-disease specific outputs
(‘HIV’ versus ‘condition’). Therefore, when partic-
ipants were asked to rate how convincing an explana-
tion was to continue to read the clinical trial in further
detail, we were implicitly measuring how important a
certain feature was for our users.
We hypothesized that search features are not
equally preferred by users. In addition, we hypoth-
esized that the formulation of explanations were not
equally preferred.
3.2.1 Results
Table 3: Features’ means and standard deviations.
Features Mean Std dev
Query in detailed description 3.69 0.82
Query in summary 3.53 0.91
Primary purpose availability 3.53 0.84
Number of publications 3.51 0.92
Stage availability 3.44 0.99
Query in title 3.15 0.93
Trial is recruiting 3.13 1
The results suggest that, in response to feature im-
portance, partial ordering can be obtained via crowd-
sourcing tasks and statistical tests. The results in Ta-
ble 3 illustrate that the feature with the highest mean
score (3.69, on a 5-point Likert scale) was Query in
detailed description, whereas the two least convinc-
ing features were Query in title, and Trial is recruit-
ing (3.15, and 3.13, respectively). We determined the
weights of our features using χ
2
tests. Table 4 pro-
vides the results of these chi-square tests where, for
example, the features Query in title and Query in sum-
mary were not equally preferred (as the results reveal
a p-value of 0.007).
Table 4: Results of feature labeling task. Note: the results
in bold are statistically significant under the assumption p
<0.05.
Features Title Summary Description Publications Stage Recruiting Primary purpose
Title / / / / / / /
Summary 0.007 / / / / / /
Description 0.00002 0.51 / / / / /
Publications 0.02 0.61 0.35 / / / /
Stage 0.038 0.67 0.15 0.88 / / /
Recruiting 0.81 0.006 0.00001 0.01 0.054 / /
Primary purpose 0.012 0.82 0.42 0.82 0.43 0.006 /
Data obtained in response to feature importance
shows that there is at least a partial ordering that can
be obtained via the crowdsourcing (based on statis-
tical tests). We determined the features’ weight, us-
ing χ
2
tests, based on statistical values. If two fea-
tures were, for example, not statistically equally pre-
ferred, these two features would be attributed differ-
ent weights.
Table 5: Experiment results for entities. Note: the results
in bold are statistically significant under the assumption p
<0.05.
Entity x(1) x(2) P-value
(1) Non-numerical
(2) Numerical
3.7 3.34 0.01
(1) Clearly mentioned
(2) Retrieved
3.65 3.33 0.036
(1) Specify disease
(2) Not specify disease
3.4 3.48 0.44
When it comes to results for the three formulation
dimensions (Table 5), when performing χ
2
tests, we
found that:
• Users prefer explanations without non-numerical
sentences (e.g. sentences mentioning that there
are ’multiple’ articles linked to the clinical trial
vs. ’2’).
• Users prefer factual sentences (‘clearly men-
tioned’) compared to actions related to the search
procedure (‘retrieved’).
• No preferences were found between specifying
the condition in a sentence (e.g. the condition
‘HIV’ was mentioned in the title) versus (‘the con-
dition’).
3.3 Explainability Score: Ordering
Retrieved Items
In this section, we used the importance of the ex-
tracted features to compute the explainability score
for each of the clinical trials and order them ac-
cordingly. To achieve this, features were assigned a
weight, which were then used to calculate the explain-
ability score and, ultimately, the clinical trials were
ordered based on these scores.
Each clinical trial was attributed an explainabil-
ity score based on its features’ availability or occur-
rences. Certain scores were fixed, while others de-
pended on the user’s query. The former are defined as
query independent features, and the latter as query de-
pendent features. As such, query dependent and query
independent features were separately calculated (Ta-
ble 2 reports which features belong to each category).
Although query dependent and query independent
scores were separately calculated, all explainability
HEALTHINF 2022 - 15th International Conference on Health Informatics
738

feature scores, shown as e
f
, were calculated in the
same manner:
e
f
= w ∗ f
s
where the explainability score for each feature is
determined by the weight w (which depends on if the
feature is binary or numeric, and if it is high or low
importance), and the feature’s score f
s
. Binary f
s
scores are identically determined for all binary fea-
tures. If the feature is present; f
s
= 1, and if the fea-
ture is unavailable; f
s
= 0. However, numerical fea-
tures f
s
scores are calculated based on each feature’s
occurrence.
As previously mentioned, the process to calculate
the scores differ for query dependent and query inde-
pendent features. For query dependent features, be-
cause all the terms related to one CUI refer to the
same condition, all e
f
scores related to one CUI were
grouped per clinical trial:
E
dtc
=
∑
e
f
dtc
where the explainability scores E for features be-
longing in the category query dependent d were calcu-
lated by grouping features’ score per CUI c for each
clinical trial t. On the other hand, features’ scores
belonging to the query independent category i scores
were calculated as:
E
it
=
∑
e
f
it
where all the E
i
t
scores are attributed to their re-
spective studies, and linked to all the study’s CUIs.
Therefore, as long as the HCP queries a condition re-
lated to the clinical trial, the E
i
t
score attributed to
that clinical trial will remain unchanged. Finally, this
score will be summed per CUI with its E
d
t
c
, which
will give us our final explainability per clinical trial
per CUI:
E
ct
= E
it
+ E
dtc
where E
c
t
was used to order the clinical trials
by conducting linear feature ranking. Explainability
scores E
c
t
range between 0 and 1, where clinical tri-
als with scores close to 1 reflect that the search en-
gine can explain more about these clinical trials com-
pared to clinical trials with a score close to 0. Hence,
the clinical trials linked to a CUI (that is queried by
the user) with the highest explainability scores for that
CUI appear higher in the results’ list.
Therefore, the algorithm orders the clinical trials
by their explainability scores e f . To do so, the algo-
rithm takes as input the HCP’s query condition. The
algorithm will then search for the query term in the
database, and identify the CUI associated to the con-
dition. Secondly, the algorithm filters all clinical trials
to keep studies related to that CUI, therefore provid-
ing a list of all articles related to the condition the
HCP queried. Lastly, the list will be ordered based on
explainability scores e f , where the highest explain-
able scores will receive the highest ordering position,
and the lowest explainability scores will receive the
lowest position.
3.4 Retrieval Explanations
Having extracted the features and computed their im-
portance, this section is dedicated on how to explain
the retrieved items in response to a user’s query. The
explanations must be written in a way that the HCP
can understand them. Therefore, we developed a tem-
plate list of sentences as shown in Table 6. These sen-
tences are simple, user-friendly, hierarchically struc-
tured, short and straightforward.
Table 6: Template sentences created for the explainability
based search engine.
Feature Template sentence
Query in title The condition is mentioned in the title
Preferred term in title The preferred term of the condition is mentioned in the title
Query in summary The condition is mentioned in the summary
Preferred term in summary The preferred term of the condition is mentioned in the summary
Query in detailed description The condition is mentioned in the detailed description
Preferred term in detailed description The preferred term of the condition is mentioned multiple times in the description
Number of publications The clinical trial has multiple publications
Stage availability The clinical trial’s stage is clearly mentioned
Overall status availability The clinical trial’s status is clearly mentioned
Trial is recruiting The clinical trial’s status is recruiting
Sentences were created in the following manner:
1. A maximum of Three Sentences at a Time Are Dis-
played. We assume that, given the limited amount
of time HCPs spend on the search engine, a max-
imum of three sentences will be enough for the
HCP to read.
2. Sentences Are Only Displayed If Certain Condi-
tions Are Met. Given the limited time this research
has, thresholds are determined based on intuitive
knowledge. This allows users to only see relevant
explanations.
3. Sentences Are Ordered by Feature Preference.
The ordering at which sentences are displayed
rely on the results of the experiment described in
sub-section 3.2.1.
4. The Sentences Are Kept Simple. To understand
which formulation of sentences users prefer, we
researched entity preferences as described in sub-
section 3.2.
4 MODEL EVALUATION
We evaluated our model by comparing it to other sim-
ulated clinical search engines based on users’ trust,
search experience, and result ordering satisfaction.
Our hypotheses were that all search engines were
Local Explanations for Clinical Search Engine Results
739

equally preferred in all three dimensions. We, there-
fore, simulated 5 different search engines with differ-
ent city names, where each engine queried either lyme
disease, breast cancer, or HIV:
1. Amsterdam: Search engine with ordered results
and with explainable sentences
2. Berlin: Search engine with ordered results and
without explainable sentences
3. Copenhagen: Search engine without ordered re-
sults and with explainable sentences
4. Dublin: Search engine without ordered results
and without explainable sentences
5. Edinburgh: Search engine with titles ordered by
alphabetical order
The engines used data from myTomorrows
2
in order
to create scenarios as realistic as possible. The dif-
ferent query concepts were queried in each search en-
gine, for which the top 10 results were extracted and
put into the simulated environments to imitate the first
page of a search engine showing 10 results at a time.
4.1 Experiment Setup
55 participants (HCPs and non-HCPs) were recruited
using different social media platforms such as Face-
book, Linkedin, or recruited in the company itself.
Participants received a link to one of the 9 question-
naires focusing on one of the query concepts (either
lyme disease, HIV, or invasive breast cancer). In each
questionnaire, participants were shown one by one the
different simulated search engines related to the query
concept. The different search engines were shown in
a random order. Additionally, participants were asked
to:
• Assess if they trusted the search engine.
– Question asked: When looking at the search en-
gine, how much do you trust the search engine?
– Possible answers: 5-point likert-scale from I
trust this engine very much to I do not trust this
search engine at all.
• Assess if they were satisfied with the ordering of
the search engines’ results
– Question asked: When looking at the search en-
gine, are you satisfied with the ordering of clin-
ical trials?
– Possible answers: 5-point likert-scale from I
am very satisfied with the ordering to I am
highly not satisfied with the ordering.
2
https://search.mytomorrows.com/public
• Assess their search experience while using the
search engine
– Question asked: What is your search experi-
ence when using the search engine?
– Possible answers: 5-point likert-scale from I
have a great search experience to My search
experience is not good at all.
In the end of the questionnaire, participants were
asked to order the different search engines by:
• Trust: users had to order the search engines from
most trustworthy to least trustworthy.
• Result Ordering Satisfaction: users were asked to
order of search engines they preferred from high-
est result ordering satisfaction to lowest result or-
dering satisfaction.
• Search Experience: users were asked to order
the search engines from best search experience to
least favourite search experience.
An example of one of the simulated search en-
gines is shown in Figure 2.
Figure 2: Example of a simulated search engine.
4.2 Results
Table 7: χ
2
results of the comparison of all search engines
to each other.
All participants HCPs Non-HCPs
Trust 0.29 0.44 0.62
Search experience 0.04∗ 0.09 0.37
Ordering satisfaction 0.31 0.40 0.48
Note: the results in bold italic with a * are statistically significant
under the assumption p <0.05. By all participants, we mean
the combination of results of HCPs and non-HCPs.
In the experiment, we asked participants to evaluate
the different search engines one by one and report
their experience. We conducted the χ
2
test to test
HEALTHINF 2022 - 15th International Conference on Health Informatics
740

our hypotheses that all search engines are equally pre-
ferred in all three dimensions. Table 7 reports the re-
sults and shows that when combining the responses
of HCPs and non-HCPs, the null hypothesis that all
search engines have equal search experience is re-
jected as the test returned a p-value of 0.04. This
suggests that users have different search experiences
when, distinctively, facing the search engines.
The results of the task asking participants to or-
der the search engines from most trusted to least
trusted, and best to worst search experience suggest
that users, both HCPs and non-HCPs, report more
trust and better search experience while using the
search engines using explainability sentences (Ams-
terdam and Copenhagen) compared to search engines
not explaining its results (Berlin, Dublin and Edin-
burgh). The results displayed in Figure 3 demonstrate
these preferences. However, when users scored the
search engines based on their ordering satisfaction,
HCPs ranked last the search engine with explainabil-
ity based ordering without explanations (Berlin) (see
Figure 4). This further demonstrates that failing to ex-
plain how the results’ ordering works leads to reduced
result ordering satisfaction.
Figure 3: Results: order the search engines from most
trusted to least trusted. Note: Results close to 1 indicate
the most preferred search engines. On the contrary, results
close to 5 are the least preferred search engines.
5 DISCUSSION
This research aimed to measure the influence of
explainability based search engines on users’ trust,
search experience and result ordering satisfaction.
Two experiments were conducted, where the first ex-
periment was created to order explainability based
features based on importance. The results were trans-
lated to weights for features to order the results re-
turned by the search engine. The second experiment
evaluated the explainability based search engine by
Figure 4: Results of ordering task: order the search engines
from best ordering satisfaction to worst ordering satisfac-
tion. Note: Results close to 1 indicate the most preferred
search engines. On the contrary, results close to 5 are the
least preferred search engines.
measuring users’ experience with the engine. Over-
all, the results suggest that search engines with expla-
nations are more trusted, provide greater user expe-
rience, and increased ordering satisfaction, compared
to search engines without explanations. In addition,
users are satisfied with explainability based ordering
of results if these have explanations, where not ex-
plaining the ordering of results decreases users’ trust,
search experience and ordering satisfaction. Thus, the
results urge developers to explain search engine re-
sults.
When asked to report the preferred order of
search engines, participants consistently preferred in
all three dimensions the search engines Amsterdam
and Copenhagen. The two search engines include
explainability based sentences and ordering, and ex-
plainability based sentences, respectively. However,
we noticed that search engine Berlin scored low, even
last in the dimension of ordering satisfaction with
HCPs. This suggests that explainability based or-
dering is preferred when explained with user-friendly
sentences. This is in line with research in (Pu and
Chen, 2006) as authors demonstrated that explainabil-
ity overall increases trust. A reason is that without ex-
plainability sentences, users understand less the logic
behind the model, and therefore attribute lower scores
to the search engine Berlin. This reasoning is further
emphasized by the significant difference in ordering
preference between Dublin and Edinburgh for non-
HCPs, where these prefer Edinburgh given that the
logic of the search engine is straightforward, which
can increase user satisfaction.
Although the model is scalable and generalizable,
the features created for this use case are not transfer-
able to other search engines. Features need to adapt
to other models’ use cases as most features created in
this research are specific to clinical trials. For exam-
ple, a search engine returning a list of travel destina-
tions would not benefit from the feature ’the clinical
Local Explanations for Clinical Search Engine Results
741

trial is recruiting’. Transferring the model as-it-is to
another set of data would, therefore, require adapt-
ing the method to the different use-case. In addition,
developers would need to collect data on feature pref-
erences for their use-case.
6 FUTURE WORK
Although our model provides explainability sen-
tences, these are not personalized to the profile of
the HCP. In order to make it more personal, results
could be ordered based on the HCP’s profile and pref-
erences. To achieve this, future work should collect
data on user profiles, and use machine learning to
identify users’ personal preferences. Moreover, this
could be combined with knowledge graphs to create
a relationship between clinical trials and users’ pro-
files as shown in (Catherine et al., 2017), where users
were provided personal explanations using a knowl-
edge graph based on item reviews and user profile.
REFERENCES
Arrieta, A. B., D
´
ıaz-Rodr
´
ıguez, N., Del Ser, J., Bennetot,
A., Tabik, S., Barbado, A., Garc
´
ıa, S., Gil-L
´
opez, S.,
Molina, D., Benjamins, R., et al. (2020). Explainable
artificial intelligence (xai): Concepts, taxonomies, op-
portunities and challenges toward responsible ai. In-
formation Fusion, 58:82–115.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1):D267–D270.
Catherine, R., Mazaitis, K., Eskenazi, M., and Cohen,
W. (2017). Explainable entity-based recommen-
dations with knowledge graphs. arXiv preprint
arXiv:1707.05254.
Das, A. and Rad, P. (2020). Opportunities and challenges
in explainable artificial intelligence (xai): A survey.
arXiv preprint arXiv:2006.11371.
Dave, D., Naik, H., Singhal, S., and Patel, P. (2020). Ex-
plainable ai meets healthcare: A study on heart dis-
ease dataset. arXiv preprint arXiv:2011.03195.
Dimanov, B., Bhatt, U., Jamnik, M., and Weller, A. (2020).
You shouldn’t trust me: Learning models which con-
ceal unfairness from multiple explanation methods. In
SafeAI@ AAAI, pages 63–73.
Dumitrache, A., Aroyo, L., and Welty, C. (2017). Crowd-
sourcing ground truth for medical relation extraction.
arXiv preprint arXiv:1701.02185.
Dumitrache, A., Aroyo, L., Welty, C., Sips, R.-J., and
Levas, A. (2013). Dr. detective”: combining gamifi-
cation techniques and crowdsourcing to create a gold
standard in medical text. In Proceedings of the 1st In-
ternational Conference on Crowdsourcing the Seman-
tic Web, volume 1030.
Gee, A. H., Garcia-Olano, D., Ghosh, J., and Paydarfar,
D. (2019). Explaining deep classification of time-
series data with learned prototypes. arXiv preprint
arXiv:1904.08935.
Lundberg, S. and Lee, S.-I. (2017). A unified approach
to interpreting model predictions. arXiv preprint
arXiv:1705.07874.
Mundhenk, T. N., Chen, B. Y., and Friedland, G. (2019). Ef-
ficient saliency maps for explainable ai. arXiv preprint
arXiv:1911.11293.
Pu, P. and Chen, L. (2006). Trust building with explanation
interfaces. In Proceedings of the 11th international
conference on Intelligent user interfaces, pages 93–
100.
Rosenfeld, A. and Richardson, A. (2019). Explainability in
human–agent systems. Autonomous Agents and Multi-
Agent Systems, 33(6):673–705.
Verma, M. and Ganguly, D. (2019). Lirme: locally inter-
pretable ranking model explanation. In Proceedings
of the 42nd International ACM SIGIR Conference on
Research and Development in Information Retrieval,
pages 1281–1284.
Zhang, Y., Mao, J., and Ai, Q. (2019). Sigir 2019 tutorial
on explainable recommendation and search. In Pro-
ceedings of the 42nd International ACM SIGIR Con-
ference on Research and Development in Information
Retrieval, pages 1417–1418.
HEALTHINF 2022 - 15th International Conference on Health Informatics
742
