
Virus Spread Modeling and Simulation: A Behavioral Parameters
Approach and Its Application to Covid-19
Alfredo Cuzzocrea
1,2,∗
and Edoardo Fadda
3,4
1
iDEA Lab, University of Calabria, Rende, Italy
2
LORIA, University of Lorraine, Nancy, France
3
DISMA, Politecnico di Torino, Torino, Italy
4
ISIRES, Torino, Italy
Keywords:
Diffusion Process, Network Dynamics.
Abstract:
How a virus spread on a network is a really important topic and even more important is to classify the danger of
a virus. With this goal in mind, we investigate the characteristics that define the most deadly virus. Moreover,
we aim to provide a simplified discrete-time simulation, described by few parameters, as a straightforward
alternative to more complex models of diseases diffusion. The simulation is used to model the spread of the
infection, and the obtained results are then analyzed to understand how the virus’ behavior varies by changing
its characteristics and the network topology.
1 INTRODUCTION
Urbanization and the destruction of natural habitats
are creating the perfect conditions for new diseases.
This problem will be increasingly frequent, as ex-
plained by Dodds (Dodds, 2019), especially due to
close contact with wildlife and livestock. Moreover,
if new diseases originates it is not easy to find a cure.
Thus, understanding how a disease spreads is the key
for prevention.
This has become clear in the last decades, from the
HIV epidemic to Ebola and, finally, with Covid-19.
Research is not only beneficial to alleviate the
workload for the health system but to improve the
overall quality of life of the population even during
a crisis, reducing the loss of human life and the long-
term effects that some diseases have on the patients’
body.
In this paper, a new approach is taken for simulat-
ing a new virus, to find what makes a virus effective
and what might help its spread. The two characteris-
tics that are analyzed are: network topology and the
dead rate and diffusion rate of the virus.
∗
This research has been made in the context of the Ex-
cellence Chair in Computer Engineering at LORIA, Nancy,
France
2 LITERATURE REVIEW
With the outbreak of coronavirus 2 (SARS-CoV-2),
many studies have been carried out to try understand-
ing the virus impact on the modern highly connected
society (e.g. (Alassafi et al., 2022; Li and Yan, 2022;
Hasaninasab and Khansari, 2022; Yadav and Vish-
wakarma, 2022; Ronaghi et al., 2022)). In the state-
of-the-art literature of epidemiological researches, the
SIR model is a widely used tool to predict the evo-
lution of infection diseases. For example, the SIQR
model, a variant of SIR model, introduces a new state
Q (Quarantine) for the individuals, and it achieves a
better health outcome for mass testing with respect to
the SIR model (Harckbart, 2020). The model SEIR,
instead, introduces a new class E for the exposed in-
dividuals who are not infectious yet, and it has been
employed, for example, to assess the effectiveness of
the policies, adopted in several Italian regions, and
their impact on future scenarios (Godio et al., 2020).
Another alternative is the SEIQR model, which in-
cludes both new states for the individuals for a more
complex and complete prediction. This last model can
be applied also to curb the impact of the transmission
of malicious objects in a highly connected computer
network (Mishra and Singh, 2011).
An alternative to the SIR models adopted in the
literature is proposed in (Kurtin et al., 2020). Here,
Cuzzocrea, A. and Fadda, E.
Virus Spread Modeling and Simulation: A Behavioral Parameters Approach and Its Application to Covid-19.
DOI: 10.5220/0011055900003179
In Proceedings of the 24th International Conference on Enterprise Information Systems (ICEIS 2022) - Volume 1, pages 187-193
ISBN: 978-989-758-569-2; ISSN: 2184-4992
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
187
the authors proposes a stochastic model to simu-
late person-to-person contact, represented as circles
bouncing in a 2D plane getting in contact with each
other.
The SIR model, with its variants, is mostly em-
ployed to simulate virus propagation and to predict
possible contagion scenarios, starting from data col-
lected on an already existing disease. In this work,
rather than predicting the virus outbreak or evaluat-
ing the effectiveness of the measures to contain the
infection, the virus is modeled in such a way that
its parameters are tuned to maximize the harmful-
ness of the virus itself, the infection and the sub-
sequent possible death. Once the optimal parame-
ters are discovered, different scenarios are considered
by either changing the virus’ parameters or varying
the network characteristics, and their impact on the
virus’ infection is analyzed. Several analogies with
the SIR model emerges throughout the analysis of the
conducted work; however, a more simplistic model
of the virus is adopted, in which, differently from
(Kurtin et al., 2020), the interactions are only pos-
sible among static neighbouring nodes. The transi-
tion among states for the individuals are not obtained
using derivatives, but they are ruled by probabilistic
conditions. In this way, the complexity of both the
virus’ behaviour and the simulation itself results to be
lower and the understanding of the overall procedure
might result more intuitive.
3 BEHAVIOUR OF THE VIRUS
There are two main characteristics that make a virus
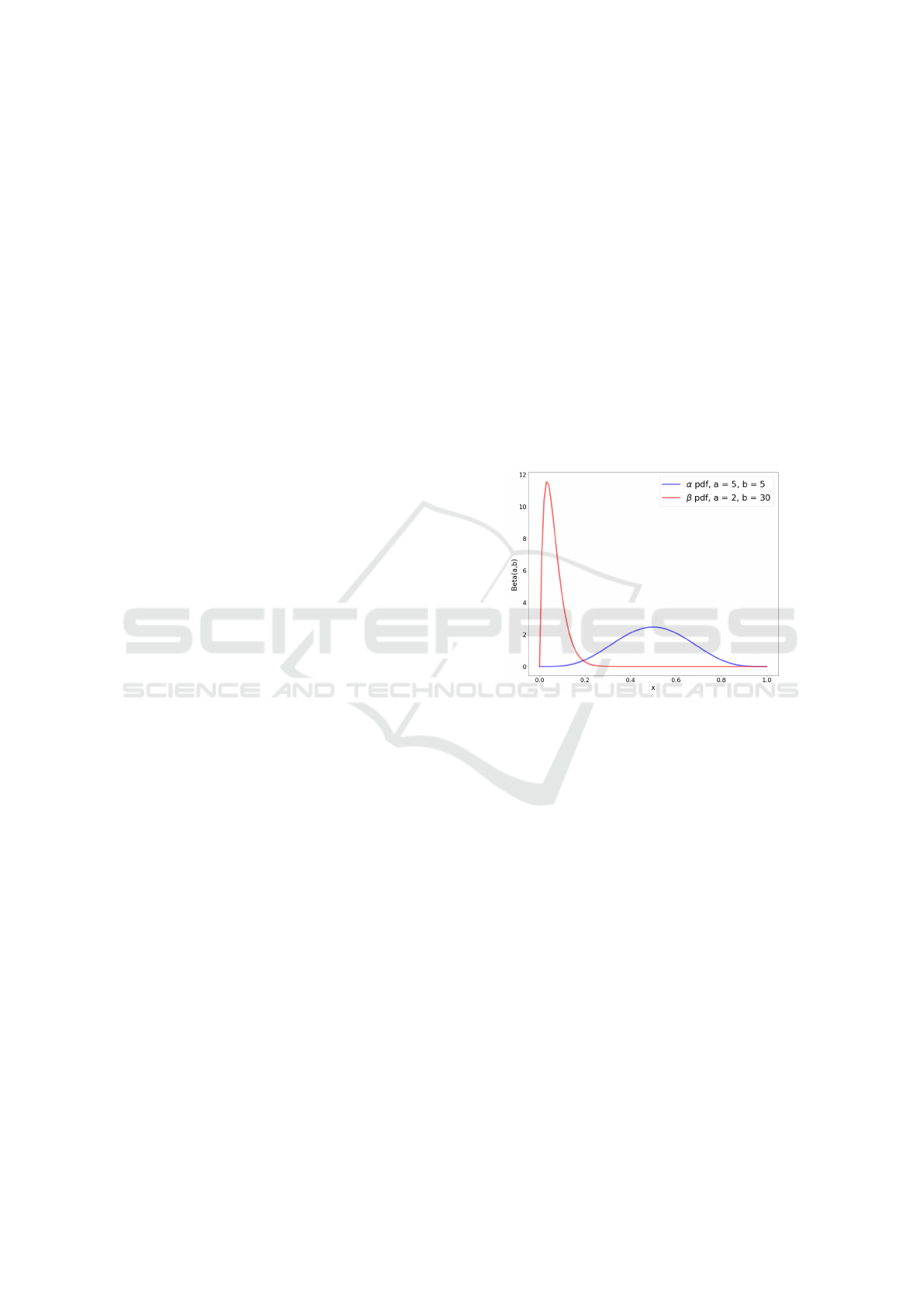
lethal: infectivity and death rate. Let us consider a
network represented by an undirected graph. Start-
ing from a random node, the infection spreads across
the network. Since the viral quantity of the virus in
the human body follows an exponential trend over
time as N
t
= N
+∞
(1 − e
−λt
), it is reasonable to as-
sume that if person i has been infected at time 0, the
probability of infecting each neighbour is defined as:
p
i
= α
i
(1 − e
−λt
) while the probability of his death is
q
i
= β
i
(1−e
−λt
), where both α
i
and β
i
are distributed
according to Beta distributions, characterized by dif-
ferent parameters (see Figure 1), and λ is the param-
eter to be optimized in order to maximize the number
of deaths. We can consider the following five states
for each node:
• Healthy: the person was not infected with the
virus;
• Contagious: the person was infected with the
virus and has a viral quantity different from zero,
therefore she/he can infect other people;
• Ill: as soon as the viral quantity exceeds the
threshold value h, 1 − e
−λt
≥ h, the person be-
comes ill and she/he is removed from the network,
in this way the quarantine period is simulated;
• Recovered: the ill person survived for T times-
tamps, hence she/he is considered healed;
• Dead: the ill person could not recover within the T
timestamps, therefore she/he is considered dead.
Figure 2 shows the flow diagram of the states a
node might go through during the simulation. Consid-
ering implementation issues, the most suitable layer
for supporting the described, flexible behaviour is rep-
resented by an XML data storage layer (e.g., (Can-
nataro et al., 2002b; Bonifati and Cuzzocrea, 2007;
Cannataro et al., 2002a; Cuzzocrea et al., 2009a)).
Figure 1: Beta distributions for both probability p to infect
someone else and probability q to die and the relative pa-
rameters.
4 THE NETWORK GRAPH
The network that has been considered is a 3000 nodes
undirected stochastic block model graph composed
by three communities. The composition of the com-
munities is defined in such a way that the three com-
munities include, respectively, 14%, 50% and 36% of
the total number of nodes. The connections among
nodes in the graph are established according to the
symmetric edge probability matrix P,
P =
0.01 0.001 0.001
0.001 0.005 0.0008
0.001 0.0008 0.005
The elements on the diagonal of matrix P de-
scribe the probability to establish connections be-
tween nodes within the same community, while the
off-diagonal elements describe the probability to con-
nect nodes belonging to different communities. Being
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
188
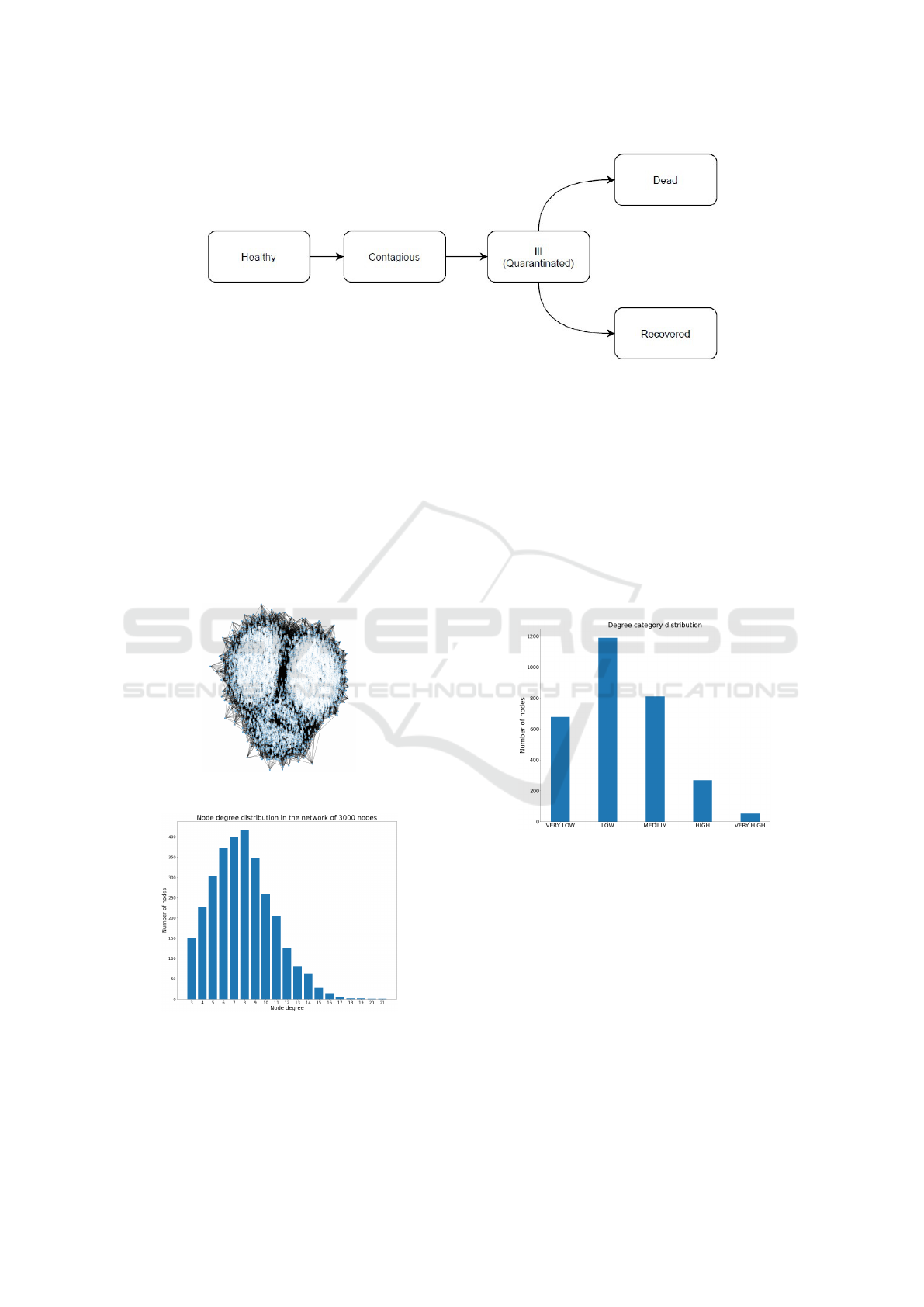
Figure 2: Flow diagram of node’s states.
the probabilities in matrix P quite small, it is likely
that some nodes will be isolated and, since patient
zero is randomly picked at the beginning of the sim-
ulation, in order to let the virus spread carry on, each
node is forced to have at least degree 3. The obtained
network has an average degree of 7.842, an average
node closeness centrality equal to 0.241 and its de-
gree distribution is shown in Figure 4. The graph is
shown in Figure 3 and it is possible to identify the
three distinct communities composing the stochastic
block model graph.
Figure 3: The actual graph of 3000 nodes.
Figure 4: Degree distribution for the stochastic block model
graph of 3000 nodes.
Each node has also been characterized by an addi-
tional information, the degree category, which identi-
fies the connectivity in broad terms of the node itself
towards the other nodes. In order to obtain this infor-
mation, the values of the node degree distribution are
subdivided into five categories. The five possible de-
gree categories are: very low, low, medium, high and
very high. By observing the degree category distri-
bution in Figure 5, it becomes clear that the vast ma-
jority of the nodes are characterized by a degree cat-
egory which is either very low, low or medium; very
few are, instead, characterized by a very high degree
category. This additional information is necessary to
observe the behaviour of the virus spread by selecting
a different degree category for the patient zero.
Figure 5: Degree category distribution of the nodes in the
graph.
5 THE SIMULATION
Having defined the graph, the parameters of the nodes
populating the graph are initialized.
More in detail, for a node i, the probability to in-
fect p
i
and the probability to die q
i
are set to zero,
since at the beginning of the simulation all the nodes
are assumed to be healthy, and the parameters α
i
and
β
i
, which rule respectively p
i
and q
i
, are drawn from
the beta distributions.
The experiment is a discrete-event simulation
which takes as input the graph with its nodes and the
Virus Spread Modeling and Simulation: A Behavioral Parameters Approach and Its Application to Covid-19
189

maximum duration in timestamps. It is also possible
to specify as input the degree category of the patient
zero or to select a specific node as patient zero. Each
timestamp of the simulation corresponds to 2 days.
5.1 Infection and Illness
In the first iteration, the patient zero node is set to be
contagious, which means that for the next iterations it
is able to infect other nodes. In the following itera-
tions, the contagious node i try to infect its neighbors.
To decide whether the neighbor node j is infected or
not by the node i, a random number is extracted from
a uniform distribution and, if it is less than p
i
, also
node j becomes contagious. Since the probability p
i
and the viral quantity are time dependent, their val-
ues are updated at each iteration for all the contagious
nodes. Sooner or later, the viral quantity of a conta-
gious node will exceed the threshold h. The occur-
rence of this event causes a state change for the node,
from the contagious state to the ill state and, from now
on, the node is forced to be in quarantine, therefore it
is not able to infect other nodes anymore. As soon as
the node becomes ill, a local counter is initialized and
incremented, iteration by iteration, for the whole ill-
ness period, and its probability to die q is initialized.
For each iteration during which the node is in the ill
state, the value of q is updated, being also its value
time dependent.
5.2 Death
Once the node i is ill, at each new iteration a random
number is drawn from a uniform distribution. If this
random number is smaller than the probability to die,
q
i
, the node is considered dead, otherwise it survives
for another iteration.
5.3 Healing
If the node is strong enough to survive the virus for a
number of iterations equal to T, the node is considered
recovered. At this point the node is immune to the
virus, it can not be infected anymore and neither it
can infect other nodes.
5.4 End of the Simulation
The simulation stops as soon as all the nodes within
the network are either healthy, healed or dead. This
scenario corresponds to not having contagious nodes
anymore, the virus spread is stopped and, in the re-
maining iterations, the ill nodes fight for their lives.
6 REAL PARAMETERS
DISCOVERY
The goal of the research is to maximize the mortality
of the virus. Since the virus behaviour is driven by
the parameters h, T and λ, by setting the values of
h and T to extremely high numbers, all the nodes of
the network will eventually die. This would make the
optimization problem trivial.
In order to model a realistic virus, the infectivity
and mortality targets are defined. The first one is set to
40%, while the second one to 7%
1
. The percentages
refer to the total amount of nodes in the network.
Several values for h and T are tested, and the pairs
of values that, jointly, get the closest to the percent-
age targets are considered. The values for λ are taken
in the interval (0,1), excluding the extremes, which
would give contagious and death probabilities that are
either null or quickly converging to α
i
and β
i
.
More in detail, the analyzed values for h, T and λ
are taken from the following ranges:
• h ∈ [0.3, 0.6], with a step of 0.05;
• T ∈ [3, 4, 5, 6];
• λ ∈ [0.05, 0.99], with a step of 0.05.
The discovery of the real (h, T ) parameters has
been carried out by selecting a patient zero node with
a degree category equal to medium. This choice pro-
vides results that are far away from the extreme sce-
narios about having either a very scarce neighborhood
or a very populated one.
For each executed simulation, characterized by a
specific triplet (h, T, λ), the discrepancy between the
actual percentage of infected nodes and the infectiv-
ity target is computed. Similarly, the discrepancy be-
tween the actual number of deaths and death target
is also discovered. The two discrepancies are then
summed up. Therefore, for each duplet of tested val-
ues (h, T ), an overall distance from the targets is ob-
tained.
Sorting the pairs (h, T ) by ascending order with
respect to the distance from the targets, only those
ones with a distance smaller than 500 are kept. The
real values for (h, T) are discovered by averaging the
obtained values. The whole list of obtained results is
shown in the following table:
The reason behind considering all the pairs (h, T )
with a distance from the targets lower than 500 relies
on the fact that, being the simulation a random exper-
iment, the same triplet (h, T, λ) could provide slightly
1
Percentages were chosen considering past pandemics,
such as the infections around the world caused by the Span-
ish influenza and the deaths for syphilis in Europe during
the XV century(Morens et al., 2020).
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
190
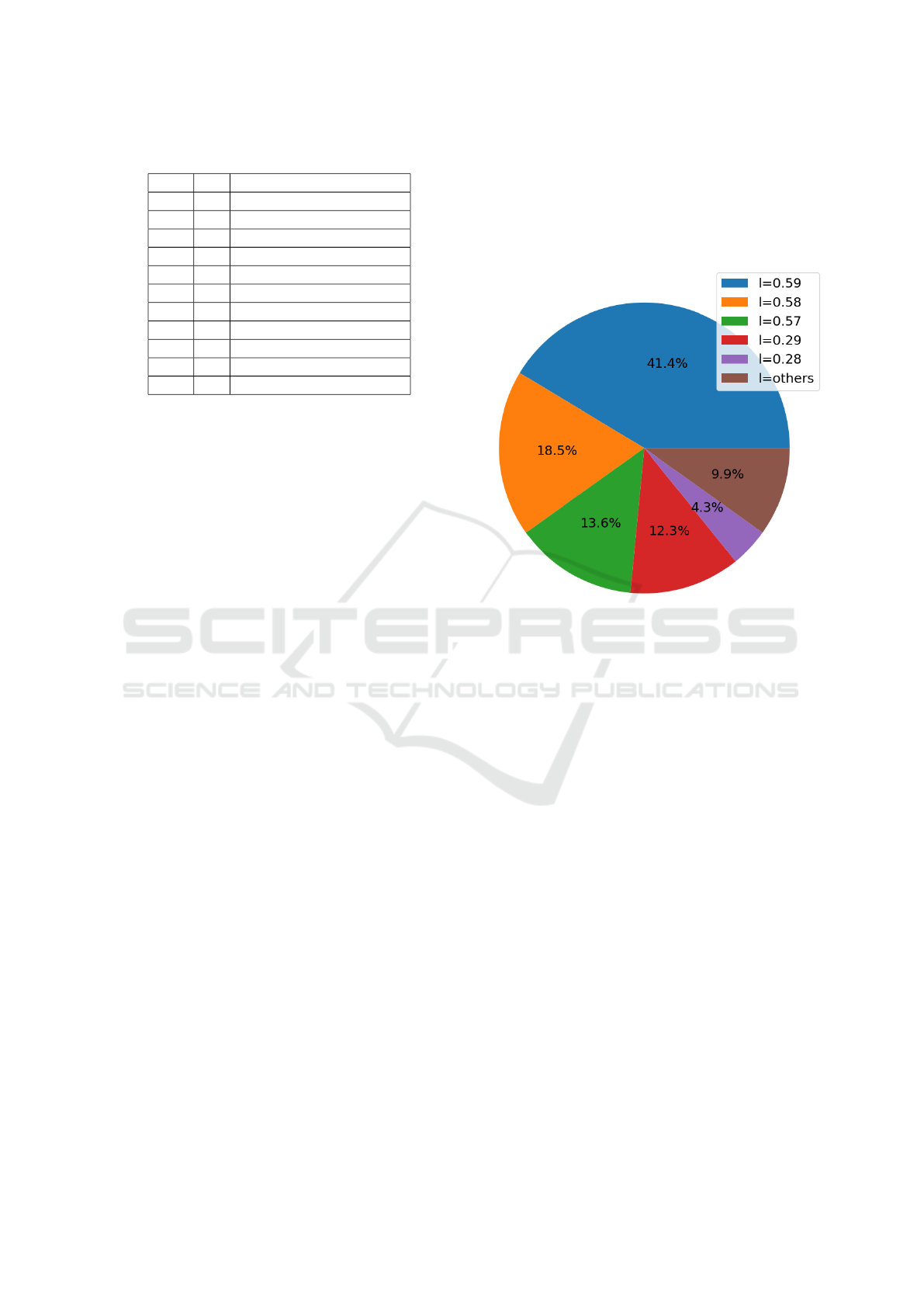
Table 1: The (h,T) pairs closest to the chosen target.
h T sum of target differences
0.3 3 17
0.45 3 136
0.4 3 176
0.35 3 229
0.55 5 342
0.45 5 372
0.3 6 374
0.3 3 432
0.5 3 434
0.3 5 446
0.55 3 487
different results over different experiments. Hence,
to alleviate this variability, a larger set of results was
considered.
The obtained real (h, T) parameters, for the net-
work of 3000 nodes, are the following:
• h = 0.45;
• T = 4.
7 LAMBDA OPTIMIZATION
In classical literature, optimization is a critical aspect
to be considered (e.g., (Cuzzocrea, 2005; Cuzzocrea
and Chakravarthy, 2010; Cuzzocrea et al., 2009b;
Cuzzocrea et al., 2003; Ceci et al., 2015)). With sim-
ilar emphasis, here we focus on how to improve sim-
ulation runs.
Now that the real (h, T ) parameters have been dis-
covered, the focus is aimed at λ and its value that
achieves the highest number of deaths in the network.
The analyzed values of λ are taken from the fol-
lowing range:
• λ ∈ [0.2, 0.7], with a step of 0.01.
So now, a more detailed range for λ is consid-
ered. The reason why the range extremes have been
reduced is given by the fact that some values of λ, ei-
ther too high or too small, provide results too far from
the virus targets previously discussed (6).
Running different simulations, the just discovered
real parameters (h, T ) are kept fixed, while λ is let
vary in the mentioned range.
The patient zero, for each simulation, has been
drawn from the list of nodes of the medium degree
category, and the number of executed simulations is
such that 20% of the nodes belonging to it are se-
lected. The reason behind this broad research is to
provide a more reliable assessment on the optimal λ
value, considering the stochastic nature of the simula-
tion.
The λ values that achieved the highest number of
deaths are reported in Figure 6.
The selected value for the optimal lambda corre-
sponds to the most recurrent lambda that achieved the
highest number of deaths. Therefore, λ
opt
= 0.59 is
the selected optimal lambda.
Figure 6: The five most recurrent lambdas that achieved the
highest number of deaths.
8 FINAL SIMULATION WITH
THE DISCOVERED
PARAMETERS
The triplet of parameters (h, T, λ), that jointly satis-
fies the infectivity and death targets and provides the
most deaths, has been discovered. A final single sim-
ulation has been executed to analyze the virus spread
evolution over time and the nodes behaviour. Figure 7
shows the results of the simulation. The patient zero
is randomly selected from the subset of nodes with a
medium degree category.
The simulation lasts 21 iterations, therefore the
virus spread takes 42 days to cease. One third of the
nodes never gets in touch with contagious nodes, and
most of the infected nodes heal from the virus, as ex-
pected. Also the total amount of ill and dead nodes
roughly respects the imposed targets; unfortunately, it
not possible to totally respect the targets, because of
the random nature of the simulation.
It can be also noticed that the temporal evolution
of the virus spread follows the SIR epidemic trend
(Keeling and Danon, 2009), in which, starting with
all the nodes being healthy, the infection begins and
Virus Spread Modeling and Simulation: A Behavioral Parameters Approach and Its Application to Covid-19
191

Figure 7: Network behaviour, patient zero of degree “MEDIUM”.
the number of ill nodes starts increasing. Once the
peak of ill nodes reaches its maximum, both recov-
ered and dead nodes increase, until a stable situation
is reached.
In order to further exploit and extend the described
simulation model, network-based solutions would be
necessary (e.g., (Cuzzocrea et al., 2005; Cuzzocrea
et al., 2004; Bellatreche et al., 2010)).
9 CONCLUSIONS AND FUTURE
WORK
The analysis carried out in this report shows that it is
possible to model a realistic virus’ behaviour, that sat-
isfies both illness and death targets, by properly tun-
ing the modelling parameters.
Furthermore, it has been shown that the network
characteristics play a fundamental role on the out-
come of the experiment. It is fair to state that, as the
overall connectivity of network increases, the virus
spreads more efficiently in the network, which might
be intuitive.
It has been also proved that the degree of the pa-
tient zero has a great impact on the development of
the virus spread, which confirms that a prompt con-
tainment of the virus is extremely effective to reduce
the virus spread, especially if the virus outbreak starts
in less connected portions of the network.
Obviously, the characteristics of the virus play a
major role. It has been discovered that the best set-
ting allows the virus to quickly spread and have a high
probability to kill the ill nodes, but not high enough
to cause the immediate death of the contagious nodes,
which would result in the death of the virus itself.
This means that, in a real case scenario, it would not
be sufficient for the health system to monitor only dis-
eases causing severe symptoms.
Finally, it may be said that the employed virus
model is quite simplistic, described by few parame-
ters, but it shows a glance of a general behaviour for
the spread of a virus, similar to the more sophisticated
SIR model.
Future work is mainly oriented to apply the de-
scribed simulation framework to innovative big data
applications (e.g., (Braun et al., 2017; Audu et al.,
2019; Ahn et al., 2019; Morris et al., 2018)).
ACKNOWLEDGEMENTS
This research has been partially supported by
the French PIA project “Lorraine Universit
´
e
d’Excellence”, reference ANR-15-IDEX-04-LUE.
The authors are grateful to R. Gallo, M. Baiocchi,
P. Migneco, M. Sangiorgio for their contribution.
REFERENCES
Ahn, S., Couture, S. V., Cuzzocrea, A., Dam, K., Grasso,
G. M., Leung, C. K., McCormick, K. L., and Wodi,
B. H. (2019). A fuzzy logic based machine learning
tool for supporting big data business analytics in com-
plex artificial intelligence environments. In FUZZ-
IEEE 2019, New Orleans, LA, USA, June 23-26, 2019,
pages 1–6. IEEE.
Alassafi, M. O., Jarrah, M., and Alotaibi, R. (2022). Time
series predicting of COVID-19 based on deep learn-
ing. Neurocomputing, 468:335–344.
Audu, A. A., Cuzzocrea, A., Leung, C. K., MacLeod, K. A.,
Ohin, N. I., and Pulgar-Vidal, N. C. (2019). An in-
telligent predictive analytics system for transportation
analytics on open data towards the development of a
smart city. In CISIS 2019, Sydney, NSW, Australia, 3-5
July 2019, pages 224–236. Springer.
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
192

Bellatreche, L., Cuzzocrea, A., and Benkrid, S. (2010).
F&A: A methodology for effectively and efficiently
designing parallel relational data warehouses on het-
erogenous database clusters. In DAWAK 2010, Bil-
bao, Spain, August/September 2010, pages 89–104.
Springer.
Bonifati, A. and Cuzzocrea, A. (2007). Efficient fragmen-
tation of large XML documents. In DEXA 2007, Re-
gensburg, Germany, September 3-7, 2007, pages 539–
550. Springer.
Braun, P., Cuzzocrea, A., Keding, T. D., Leung, C. K., Paz-
dor, A. G. M., and Sayson, D. (2017). Game data
mining: Clustering and visualization of online game
data in cyber-physical worlds. In KES 2017, Mar-
seille, France, 6-8 September 2017, pages 2259–2268.
Elsevier.
Cannataro, M., Cuzzocrea, A., Mastroianni, C., Ortale, R.,
and Pugliese, A. (2002a). Modeling adaptive hyper-
media with an object-oriented approach and XML. In
WebDyn@WWW 2002, Honululu, HW, USA, May 7,
2002, pages 35–44. CEUR-WS.org.
Cannataro, M., Cuzzocrea, A., and Pugliese, A. (2002b).
XAHM: an adaptive hypermedia model based on
XML. In SEKE 2002, Ischia, Italy, July 15-19, 2002,
pages 627–634. ACM.
Ceci, M., Cuzzocrea, A., and Malerba, D. (2015). Ef-
fectively and efficiently supporting roll-up and drill-
down OLAP operations over continuous dimensions
via hierarchical clustering. J. Intell. Inf. Syst.,
44(3):309–333.
Cuzzocrea, A. (2005). Overcoming limitations of approx-
imate query answering in OLAP. In IDEAS 2005,
25-27 July 2005, Montreal, Canada, pages 200–209.
IEEE Computer Society.
Cuzzocrea, A. and Chakravarthy, S. (2010). Event-based
lossy compression for effective and efficient OLAP
over data streams. Data Knowl. Eng., 69(7):678–708.
Cuzzocrea, A., Darmont, J., and Mahboubi, H. (2009a).
Fragmenting very large XML data warehouses via k-
means clustering algorithm. Int. J. Bus. Intell. Data
Min., 4(3/4):301–328.
Cuzzocrea, A., Furfaro, F., Greco, S., Masciari, E., Mazzeo,
G. M., and Sacc
`
a, D. (2005). A distributed system
for answering range queries on sensor network data.
In PerCom 2005 Workshops, 8-12 March 2005, Kauai
Island, HI, USA, pages 369–373. IEEE Computer So-
ciety.
Cuzzocrea, A., Furfaro, F., Masciari, E., Sacc
`
a, D., and Sir-
angelo, C. (2004). Approximate query answering on
sensor network data streams. GeoSensor Networks,
49.
Cuzzocrea, A., Furfaro, F., and Sacc
`
a, D. (2003). Hand-
olap: A system for delivering OLAP services on hand-
held devices. In ISADS 2003, 9-11 April 2003, Pisa,
Italy, pages 80–87. IEEE Computer Society.
Cuzzocrea, A., Furfaro, F., and Sacc
`
a, D. (2009b). En-
abling OLAP in mobile environments via intelligent
data cube compression techniques. J. Intell. Inf. Syst.,
33(2):95–143.
Dodds, W. (2019). Disease now and potential future pan-
demics. The World’s Worst Problems, page 31–44.
Godio, A., Pace, F., and Vergnano, A. (2020). Seir model-
ing of the italian epidemic of sars-cov-2 using compu-
tational swarm intelligence. IJERPH, 17.
Harckbart, G. (2020). Heterogeneous siqr models with mass
testing and targeted quarantine and the spread of in-
fectious disease.
Hasaninasab, M. and Khansari, M. (2022). Efficient
COVID-19 testing via contextual model based com-
pressive sensing. Pattern Recognit., 122:108253.
Keeling, M. J. and Danon, L. (2009). Mathematical mod-
elling of infectious diseases. British Medical Bulletin,
92(1):33–42.
Kurtin, D. L., A.J., D., and Stagg, S. M. (2020). VTES:
a stochastic python-based tool to simulate viral trans-
mission [version 1]. F1000Research.
Li, S. and Yan, Y. (2022). Data-driven shock impact of
COVID-19 on the market financial system. Inf. Pro-
cess. Manag., 59(1):102768.
Mishra, B. K. and Singh, A. K. (2011). Two quarantine
models on the attack of malicious objects in computer
network. Hindawi Publishing Corporation.
Morens, D. M., Daszak, P., Markel, H., and Taubenberger,
J. K. (2020). Pandemic covid-19 joins history’s pan-
demic legion. mBio, 3(11).
Morris, K. J., Egan, S. D., Linsangan, J. L., Leung, C. K.,
Cuzzocrea, A., and Hoi, C. S. H. (2018). Token-based
adaptive time-series prediction by ensembling linear
and non-linear estimators: A machine learning ap-
proach for predictive analytics on big stock data. In
ICMLA 2018, Orlando, FL, USA, December 17-20,
2018, pages 1486–1491. IEEE.
Ronaghi, F., Salimibeni, M., Naderkhani, F., and Mo-
hammadi, A. (2022). COVID19-HPSMP: COVID-19
adopted hybrid and parallel deep information fusion
framework for stock price movement prediction. Ex-
pert Syst. Appl., 187:115879.
Yadav, A. and Vishwakarma, D. K. (2022). A language-
independent network to analyze the impact of
COVID-19 on the world via sentiment analysis. ACM
Trans. Internet Techn., 22(1):28:1–28:30.
Virus Spread Modeling and Simulation: A Behavioral Parameters Approach and Its Application to Covid-19
193
