
Sensitive Detection of BRAF Hotspot Mutation V600E using
E-ice-COLD-PCR Combined with Pyrosequencing
Fei Yu
a
, Kunxian Shu
b
and Dan Pu
c
Chongqing Key Laboratory of Big Data for Bio Intelligence, Chongqing University of Posts and Telecommunications,
Chongqing, China
Keywords: E-ice-COLD-PCR, Pyrosequencing, BRAF, V600E.
Abstract: It is important to sensitively detect BRAF V600E mutation, since it is an important biological marker in several
types of cancers, such as melanomas and colorectal cancers. Here, we have combined enhanced improved and
complete enrichment co-amplification at lower denaturation temperature-polymerase chain reaction (E-ice-
COLD-PCR) with pyrosequencing for the detection of BRAF V600E mutation. A serial of mutation-
containing dilutions was determined, and the detect limit of E-ice-COLD-PCR/pyrosequencing and
conventional PCR/pyrosequencing assays were 0.1 and 5%, respectively. When BRAF V600E mutation in 10
melanoma patients were further determined, all samples with the V600E mutation detected by conventional
PCR/pyrosequencing were also found to be detectable with E-ice-COLD-PCR/pyrosequencing. However, one
sample was detected by using E-ice-COLD-PCR/pyrosequencing but not by conventional
PCR/pyrosequencing. In summary, E-ice-COLD-PCR/pyrosequencing is sensitive, reliable and cost-effective
for detecting BRAF V600E mutation in clinical samples.
1 INTRODUCTION
1
Mutations in BRAF have been found in a wide range
of human cancers, including melanomas (59%),
colorectal cancers (10%), thyroid cancers (30–70%),
and early ovary cancers (30%) (Besaratinia 2008). Of
those mutations, BRAF V600E mutation which
replaces the valine by glutamate at codon 600 is the
most frequent mutation, accounting for ~92% of all
BRAF cancer mutations (Besaratinia 2008). Thus, it
is clinically important to detect BRAF V600E
mutation. Sanger sequencing is the gold standard for
molecular diagnosis, and it is accurate and reliable
(How-Kit 2013). However, it has several limitations
in terms of costs effectiveness and sensitivity. For
example, its limit of detection is only 10–30% of
mutated alleles in a background of wild-type alleles
(How-Kit 2013). Mostly, the DNA abundance or
concentration of specimen samples obtained from
tumor tissues and liquid biopsies, such as cell-free
DNA (cfDNA), may be lower than 1%. In this case,
the low sensitivity of Sanger sequencing will not be
a
https://orcid.org/0000-0003-3069-0156
b
https://orcid.org/0000-0003-4487-2727
c
https://orcid.org/0000-0002-0156-898X
sufficient to detect the aforementioned low-
abundance BRAF V600E mutation.
Pyrosequencing is a real-time DNA sequencing
technique which measures the sensitivities of emitted
light during DNA synthesis. It employs a set of
enzymatic reactions to generate inorganic PPi and to
convert it to visible light during the polymerization.
This technology adds one nucleotide into the reaction
at a time during DNA synthesis. Due to the previously
known on the type of bases added, the sequence of the
determined template can be interrogated sequentially
(Tan 2008). Pyrosequencing offers a specific,
sensitive, rapid and cost-effective alternative to
dideoxy sequencing for the detection of BRAF V600E
mutation (Tan 2008). Although pyrosequencing has a
low detection limit of 5%, it still does not enable to
detect the low percentage of mutated DNA.
To further decrease the limit of mutation
detection, methods which are capable of enriching the
unknown mutations in samples have been developed.
For example, Co-amplification at lower denaturation
temperature PCR (COLD-PCR) has been established
Yu, F., Shu, K. and Pu, D.
Sensitive Detection of BRAF Hotspot Mutation V600E using E-ice- COLD-PCR Combined with Pyrosequencing.
DOI: 10.5220/0011230300003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 535-539
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
535

to highly enrich low-abundance mutations from
clinical samples, and the detection efficiency is
independent of the mutation type and position (Li
2008). However, COLD-PCR is not able to
effectively enrich and identify all forms of unknown
mutations (Li 2008, Milbury 2011). Another
drawback of COLD-PCR is that it needs to very
precisely control a critical temperature (T
c
) as a slight
variation of 0.2°C may completely fail the mutation
enrichment (Milbury 2011). Therefore, by the
inclusive of a wild-type (WT) specific blocker probe,
an improved and complete enrichment COLD-PCR
(ice-COLD-PCR), in which a WT specific blocker
probe was included, has been proposed to eliminate
shortcoming of COLD-PCR that cannot enrich all
mutation types (Milbury 2011). Further, by the
incorporation of locked–nucleic acid (LNA)
nucleotides in the WT blocker probe, a relatively
novel technology, Enhanced-ice-COLD-PCR has
also been developed. It maximizes the T
m
differences
between homo- and heteroduplexes for a single base
mis-match and thus overcomes the above-mentioned
issue that acquires a critical T
c
in the COLD-PCR
assays (How-Kit 2013). This technique provides a
sensitive, reproducible and flexible technique for
mutation enrichment.
In the present study, we demonstrate that E-ice-
COLD-PCR is used to enrich low abundance BRAF
V600E mutation and pyrosequencing is then applied
as the downstream readout technology. The
efficiency of E-ice-COLD-PCR/pyrosequencing is
evaluated in detecting BRAF hotspot mutation
V600E.
2 MATERIALS AND METHODS
2.1 Samples and DNA Extraction
Tumor specimens were obtained from 10 melanoma
patients in this study. Genomic DNA from formalin-
fixed paraffin-embedded (FFPE) tissue was extracted
using the QIAamp DNA FFPE Tissue Kit (Qiagen)
following the manufacturer’s instructions. After
extraction and purification, the DNA was quantified
using a Qubit fluorometer (Thermo Fisher, Waltham,
MA).
2.2 Plasmid Standards
Plasmid DNA templates harbored BRAF V600E
mutation in exon 15 of BRAF gene were generated. A
plasmid containing wild-type BRAF exon 15 was also
constructed. The mutation type of plasmids was
verified by using a PyroMark Q96 MD
pyrosequencing instrument (Qiagen, Courtaboeuf,
France). All the plasmids of the defined genotypes
were used to generate amplicons for positive and
negative controls.
2.3 Conventional PCR
Twenty-five nanogram of genomic DNA was used as
template in a 25-μL PCR mix including 1× HotStar
Taq DNA polymerase buffer, 1.6 mM of additional
MgCl
2
, 200 μM of dNTPs, 200 nM of the forward and
reverse primers, and 2 U of HotStar Taq DNA
polymerase. PCR cycling conditions contained an
initial 95◦C denaturation for 3 min, followed by 40
cycles of 95◦C for 30 s, 56◦C for 30 s, 72◦C for 10 s,
and 72°C for 5 min. Amplicons were verified by gel
electrophoresis on a 2% agarose gel prior to
pyrosequencing.
2.4 E-ice-COLD-PCR
E-ice-COLD-PCR reaction volume was 50 µL and
the reaction mixtures contained 1× of Phusion DNA
polymerase buffer, 25 ng of genomic DNA, 200 μM
of dNTPs, 200 nM of each primer, 1.6 mM of
additional MgCl
2
, 40 nM of the blocker probe, and 5
U of Phusion
TM
DNA polymerase (New England
Biolabs (NEB)). E-ice-COLD-PCR protocol
contained an initial 95◦C denaturation for 10 min,
followed by 6 cycles of 95◦C for 30 s, 60◦C for 20 s,
72◦C for 10 s, and then 44 cycles of 95◦C for 20 s,
70◦C for 30 s, 60◦C for 20 s, 72◦C for 10 s, and a final
extension at 72°C for 5 min.
2.5 Pyrosequencing
Twenty µL of conventional PCR or E-ice-COLD-
PCR products were purified and the purified products
were applied to generate single-stranded for
sequencing reaction by use of a PyroMark Q96
Vacuum Workstation (Qiagen, Courtaboeuf, France).
Thereafter, the sequencing primer was annealed to the
single-stranded target sequence after incubation at
80°C for 2 min. The order of nucleotide dispensation
was C/A/G/A/T/G/A/T/C. After the run, the
abundance of each genotype was determined by the
AQ model of Pyromark Q96 MA software and shown
upon the pyrogram.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
536
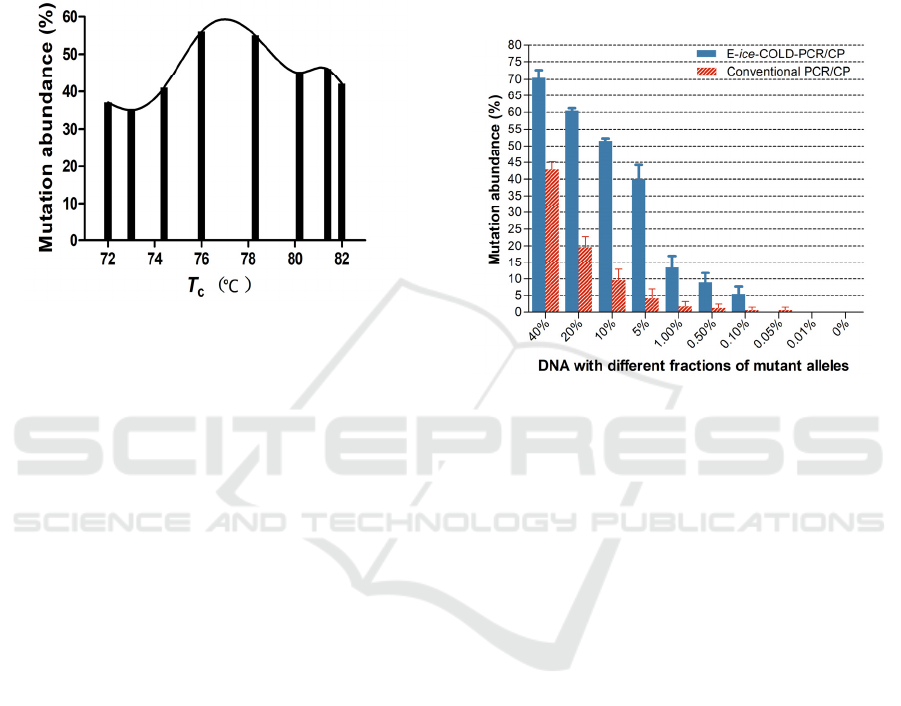
3 RESULTS
3.1 The Determination of T
c
It is important to determinate the optimal critical T
c
for E-ice-COLD-PCR reactions, at which mutant
sequences were robustly amplified as well as
Figure 1: Determination of suitable critical denaturation
temperatures (Tc) using a set of E-ice-COLD-PCR assays
with gradient Tc (from 72◦C to 82◦C) on 5% mutation
fraction in mixed plasmids and 40 nM of blockers.
at 5% mutant abundance with 40 nM of blocker
probes. The PCR products were analyzed by
pyrosequencing. As shown in Figure 1, with the
increase of the Tc, the enrichment efficiency of
mutant sequences increased. When Tc was 76°C,
mutant sequences could be effectively enriched.
However, when the Tc is higher than 78.3°C, the
enrichment efficiency of mutant sequences gradually
decreased. Thus, 76°C was chosen as the appropriate
T
c
for the E-ice-COLD-PCR in this study.
3.2 Comparison of Standard
PCR/Pyrosequencing and
E-ice-COLD-PCR/Pyrosequencing
for V600E Mutation Detection
To compare conventional PCR/pyrosequencing and
E-ice-COLD-PCR/pyrosequencing in detecting low-
level BRAF V600E mutation, plasmid DNA with
BRAF V600E mutation was diluted serially into wild-
type DNA to generate the following fractions of
mutations: 40, 20, 10, 5, 1, 0.5, 0.1, 0.05, 0.01 and
0%. pyrosequencing (Figure 2). As shown, the
mutation abundances obtained by both conventional
PCR/pyrosequencing and E-ice-COLD-
PCR/pyrosequencing decreased, as the ratios of the
mutant to wild-type plasmids decreased. Since the
reported detect limit of pyrosequencing was
approximately 5% (Mauger 2017), mutation
abundance lower than 5% was undetectable. Thus, E-
ice-COLD-PCR/pyrosequencing was capable of
detecting BRAF V600E mutation at a much lower
ratio of mutant to wild-type alleles (0.1%) than
conventional PCR/pyrosequencing (5%). That was to
say, the detection limit of pyrosequencing was as low
as 0.1% when combined with upstream enrichment E-
ice-COLD-PCR.
Figure 2: Comparison of conventional PCR/pyrosequencing
and E-ice-COLD-PCR/pyrosequencing for BRAF V600E
mutation detection.
3.3 E-ice-COLD-PCR/Pyrosequencing
for BRAF V600E Detection in
Clinical Samples
To further explore the potential clinical applications of
E-ice-COLD-PCR/pyrosequencing for BRAF V600E
detection in clinical samples, FFPE tissues from 10
melanoma patients were enriched by E-ice- COLD-
PCR followed by pyrosequencing. The same samples
were also analyzed by using conventional
PCR/pyrosequencing for comparison. As shown in
Table 1, the V600E mutations in all the five samples
that were positive by conventional
PCR/pyrosequencing were successfully detected by
using E-ice-COLD-PCR/pyrosequencing.
Furthermore, among the 5 undetectable samples, one
of them with no V600E mutation detected by
conventional PCR/pyrosequencing was successfully
detected by E-ice-COLD-PCR/pyrosequencing. That
was, E-ice-COLD-PCR/pyrosequencing increased the
mutations detected in clinical melanoma specimens by
10% (1/10). Therefore, E-ice-COLD-
PCR/pyrosequencing is more sensitive in detecting the
V600E mutation when compare with conventional
PCR/pyrosequencing, which might assist in cancer
treatment and monitoring and in prenatal diagnosis.
Sensitive Detection of BRAF Hotspot Mutation V600E using E-ice- COLD-PCR Combined with Pyrosequencing
537

Table 1: Comparison of conventional PCR/pyrosequencing and E-ice-COLD-PCR/pyrosequencing assays for BRAF V600E
mutation detection in clinical samples.
No. Mutation Conventional PCR/pyrosequencing E-ice-COLD-PCR/pyrosequencing
S1 c.1799T > A WT
a
WT
S2 c.1799T > A WT WT
S3 c.1799T > A WT
MT
S4 c.1799T > A MT
b
MT
S5 c.1799T > A MT MT
S6 c.1799T > A MT MT
S7 c.1799T > A WT WT
S8 c.1799T > A MT MT
S9 c.1799T > A WT WT
S10 c.1799T > A MT MT
The grey highlighted sample was with no V600E mutation detected by conventional PCR/pyrosequencing but successfully
detected by E-ice-COLD-PCR/pyrosequencing. a: Wild-type (WT). b: Mutant-type (MT).
4 DISCUSSIONS
With the introduction of precision medicine, the
ability to detect low-abundance mutation or DNA has
become more and more important in several clinical
areas including diagnosis, treatment and prognosis of
cancers, non-invasive prenatal diagnosis, forensic
identification and so on. Here, we combined E-ice-
COLD-PCR with pyrosequencing to detect BRAF
V600E mutation in serial dilutions of mutant DNA
and in clinical samples. The results demonstrated that
E-ice-COLD-PCR/pyrosequencing showed higher
levels of enrichment and sensitivity over
conventional PCR/pyrosequencing. The detect limit
of 0.1% in this work was consistent with the reported
work for KRAS mutations detection (How-Kit 2013).
However, it was slightly lower than that of the
previously published work in which an E-ice-COLD-
PCR/pyrosequencing assay has been used for
detecting BRAF mutations with a detection limit of
0.01% (How-Kit 2014). The main reason might be
that the different concentrations of blocker probes for
different types of samples or different amounts of
DNA input might show different detection quality
and quantity (How-Kit 2014).
The widely used downstream read-out
technologies includes HRM analysis, Sanger
sequencing, and NGS. These methods are either not
sensitive enough or too expensive when used in
detecting low-abundance BRAF V600E mutation.
Pyrosequencing is relatively sensitive (with a
detection limit of 5%) and cost-effective for mutation
detection. It enables to detect 0.1–0.01% of mutant
DNA, when combined with E-ice-COLD-PCR to
analyze mutations (How-Kit 2013, How-Kit 2014). In
this study, the samples were analyzed in duplicates. A
5% mutation threshold for pyrosequencing was
considered a simple as mutant type corresponding to
the limit of detection of pyrosequencing. The samples
were considered as mutated if their abundances
showed a mutation level higher than 5%. In addition,
when FFPE tissues were analyzed by E-ice-COLD-
PCR/pyrosequencing, one sample with no mutation
detected by conventional PCR/pyrosequencing was
successfully detected by using E-ice-COLD-
PCR/pyrosequencing. The results clearly
demonstrated the power of E-ice-COLD-
PCR/pyrosequencing in BRAF V600E mutation
detection and identification in clinical samples.
5 CONCLUSIONS
All in all, we have combined E-ice-COLD-PCR with
pyrosequencing to detect BRAF hotspot mutation
V600E, and demonstrated that the use of E-ice-
COLD-PCR/pyrosequencing increased the sensitivity
for detecting BRAF V600E mutation with detection
limit of 0.1%. When FFPE tissue specimens were
detected, one sample was detected by using E-ice-
COLD-PCR/pyrosequencing but not by conventional
PCR/pyrosequencing. Thus, E-ice-COLD-
PCR/pyrosequencing is high sensitivity, reproducible
and flexible for identifying low-abundance BRAF
V600E mutation in clinical specimens.
ACKNOWLEDGEMENTS
The work was funded by the National Science
Foundation of China [61801071], and the Basic
Research and Frontier Exploration Project of
Chongqing Science and Technology Commission
[CSTC2018jcyjAX0246].
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
538

REFERENCES
Besaratinia, A., Pfeifer, G.P. (2008) Sunlight ultraviolet
irradiation and BRAF V600 mutagenesis in human
melanoma. Hum. Mutat. 29, 983–991.
How-Kit, A., Mazaleyrat, N., Daunay, A., Nielsen, H.M.,
Terris, B., Tost, J. (2013) Sensitive detection of KRAS
mutations using enhanced-ice-COLD-PCR mutation
enrichment and direct sequence identification. Hum.
Mutat. 34,1568–1580.
How-Kit, A., Lebbe, C., Bousard, A., Daunay, A.,
Mazaleyrat, N., Daviaud, C., Mourah, S., Tost, J.
(2014) Ultrasensitive detection and identification of
BRAF V600 mutations in fresh frozen, FFPE, and
plasma samples of melanoma patients by E-ice-COLD-
PCR. Anal. Bioanal. Chem., 406: 5513–5520.
Li, J., Wang, L., Mamon, H., Kulke, M.H., Berbeco, R.,
Makrigiorgos, G.M. (2008) Replacing PCR with
COLD-PCR enriches variant DNA sequences and
redefines the sensitivity of genetic testing. Nat. Med.
14, 579–584.
Milbury, C.A., Li, J., Makrigiorgos, G.M. (2011) Ice-
COLD-PCR enables rapid amplification and robust
enrichment for low-abundance unknown DNA
mutations. Nucleic Acids Res. 39, e2.
Mauger, F., How-Kit, A., Tost, J. (2017) COLD-PCR
Technologies in the Area of Personalized Medicine:
Methodology and Applications. Mol Diagn Ther. 21(3),
269-283.
Tan, Y.H., Liu, Y., Eu, K.W., Ang, P.W., Li, W.Q., Salto-
Tellez, M., Iacopetta, B., Soong, R. (2008) Detection of
BRAF V600E mutation by pyrosequencing. Pathology.
40, 295–298.
Sensitive Detection of BRAF Hotspot Mutation V600E using E-ice- COLD-PCR Combined with Pyrosequencing
539
