
Networks and Algorithms in Heterogeneous Network-based Methods
for Drug-target Interaction Prediction: A Survey and Comparison
Shanglin Gao
1a
, Zhixing Liu
2,* b
and Ying Hong Li
1,* c
1
Chongqing Key Laboratory of Big Data for Bio Intelligence, Chongqing University of Posts and Telecommunications,
Chongqing, China
2
The Second People's Hospital of Jiulongpo District, Chongqing, China
*
Corresponding author
Keywords: Drug-Target Interaction, Heterogeneous Network-Based Methods, Networks, Algorithms.
Abstract: A key step in drug discovery is the identification of drug-target interactions (DTIs). However, only a small
fraction of DTIs have been experimentally validated due to the time-consuming and expensive aspects of
experimental validation. To improve the efficiency of drug discovery, many computer-aided drug-target
prediction methods have been developed to guide experimental validation. There are numerous prediction
methods for DTIs, among which heterogeneous network-based methods do not depend on the 3D structures
of the targets or compound molecules and they avoid the shortcomings of machine learning methods for
negative training dataset selection, exhibiting greater advantages than other methods. Currently, although
many reviews of drug-target prediction methods exist, only a few of them have addressed network-based
methods, and they have not been compared in terms of the heterogeneous networks and algorithms used.
Therefore, this paper presents a review of the heterogeneous network-based methods for DTI prediction,
compares the differences in the prediction performance of different heterogeneous networks and algorithms
from the perspective of the networks and algorithms used by these methods, and provides suggestions for
the selection of heterogeneous networks and algorithms.
1 INTRODUCTION
1
Drug-target interactions (DTIs) can be
experimentally validated by wet-laboratory methods
(e.g., affinity chromatography, etc.) (Bi et al. 2015).
However, these experiments are time-consuming
and costly, and large-scale validation is not possible.
Therefore, predicting DTIs by computer-assisted
methods will significantly reduce the scope of
experimental validation and improve the efficiency
of drug discovery. With the rapid increase in the
number of compounds (Kim et al. 2021), the
proportion of compound molecules with known
target characteristics and drug effects has decreased.
In addition, researchers have accumulated a large
amount of information on compounds, proteins, and
interactions to construct larger datasets, making it
a
https://orcid.org/0000-0003-4735-7245
b
https://orcid.org/0000-0002-1351-9149
c
https://orcid.org/0000-0003-3629-519X
possible to develop more accurate and efficient
methods to predict DTIs.
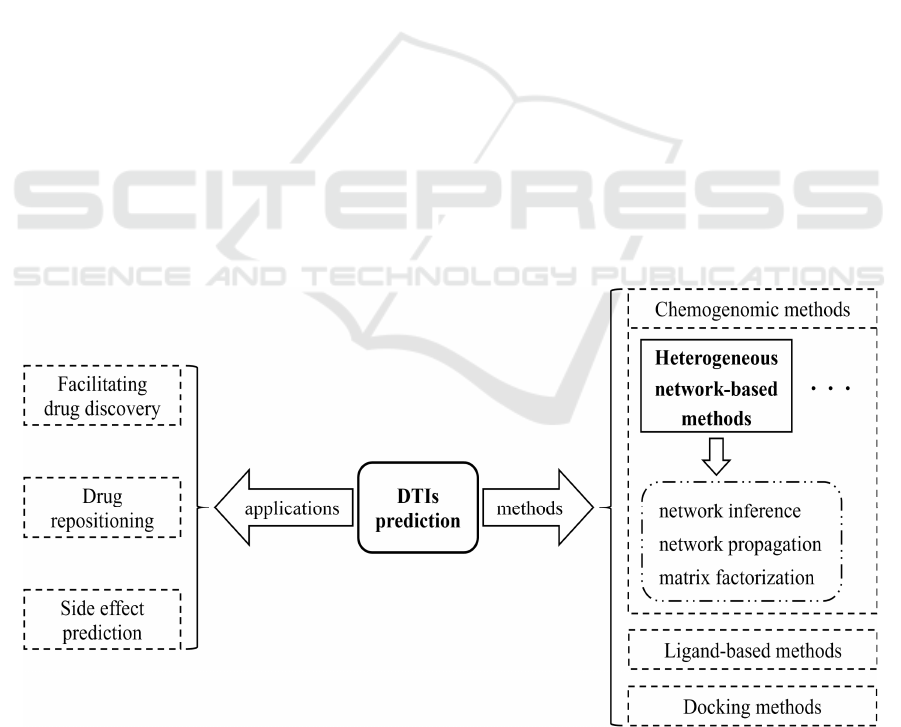
DTI prediction has multiple applications, such as
facilitating drug discovery (Chen Z. H. et al. 2020),
drug repositioning (Chen Z. H. et al. 2020), and drug
side-effect prediction (Pliakos & Vens 2020). The
drug discovery process is long, has a low success
rate, and consumes significant resources. It is
estimated that it takes approximately 10–15 years to
develop a new drug, consuming an average of $1.8
billion (Paul et al. 2010). Currently, the main reason
why the vast majority of the compounds that have
been discovered are not used as drugs is that the
interaction of these compounds with proteins is
unknown. Therefore, a computer-aided approach to
predict compound-protein interactions would have
the potential to significantly narrow the drug search
space and improve the efficiency of drug discovery.
Drug repositioning is a research strategy for new
uses outside the scope of the original medical
indication for a marketed drug or a clinical trial drug
(Ashburn & Thor 2004). The safety of approved
Gao, S., Liu, Z. and Li, Y.
Networks and Algorithms in Heterogeneous Network-based Methods for Drug-target Interaction Prediction: A Survey and Comparison.
DOI: 10.5220/0011230500003438
In Proceedings of the 1st International Conference on Health Big Data and Intelligent Healthcare (ICHIH 2022), pages 67-75
ISBN: 978-989-758-596-8
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
67
drugs or clinical trial drugs has been widely
confirmed due to the extensive clinical trials they
have undergone. Since the outbreak of the COVID-
19 pandemic, drug repositioning has become a
method for the rapid development of potent anti-
COVID-19 drugs. Drug repositioning studies can
either directly predict drug molecules for treating a
disease or to screen potential drug molecules by DTI
prediction in the context of identifying therapeutic
targets. DTI prediction has become an important
research direction in drug repositioning. The
combination of a drug with a therapeutic target may
produce therapeutic effects, while the combination
of a drug with other targets may produce side
effects. Drug side effects have become a major cause
of drug clinical trial failure (Pliakos & Vens 2020).
Therefore, predicting possible drug side effects by
DTI at the preclinical study stage will help in
selecting more suitable drug molecules for clinical
trials.
Therefore, DTI prediction will be very useful in
drug development. Prediction methods for DTIs are
generally divided into three categories (Sachdev &
Gupta 2019): ligand-based methods, docking
methods, and chemical genomics methods. Ligand-
based methods were developed based on the idea
that similar molecules usually bind to similar protein
targets and display similar properties (Jacob & Vert
2008). Docking methods use simulations of the
three-dimensional structures of proteins and drugs to
predict whether they will interact with each other
(Nagamine et al. 2009). Chemogenomic approaches
use information from both drugs and proteins for
interaction predictions (Zhao et al. 2019).
Heterogeneous network-based methods are the
best type of chemical genomics methods for
prediction (Ezzat et al. 2019), which do not depend
on the 3D structure of targets and compound
molecules or avoid the defects of negative data
selection of machine learning methods, showing
greater advantages than other methods. The methods
based on heterogeneous networks can be generally
classified into network inference (Saint-Antoine &
Singh 2020; Cheng et al. 2012), network
propagation (Engin et al. 2014), and matrix
decomposition (Hodos et al. 2016; Abbou et al.
2021) (Fig. 1). Several review articles have been
published about the prediction methods for DTIs,
which also contain a summary of the network-based
prediction methods (Wu et al. 2018). However, these
reviews do not provide a systematic comparison of
the heterogeneous networks and algorithms used by
these prediction algorithms. Therefore, this thesis
reviews recent heterogeneous network-based
forecasting methods for DTIs and proposes
recommendations for heterogeneous network
construction and algorithm selection after a
systematic comparison of the heterogeneous
networks and algorithms used.
Figure 1: The application and classification of DTIs.
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
68

2 DTIs PREDICTIONS METHODS
This review summarizes the newly published
prediction methods for DTIs in recent years and
classifies them into the following categories:
network propagation, network inference, and matrix
factorization (Table 1).
Network propagation is a common approach
used to analyze heterogeneous networks, and a
variety of DTI prediction tools have been developed
based on this approach. NRWRH (Chen et al. 2012)
is a large-scale method for predicting DTIs
constructed by Chen et al. using a restarting random
walk algorithm under the assumption that similar
drugs usually have similar targets. This method
integrates three different networks (a protein
similarity network, a drug similarity network, and a
drug-target interaction network) into a “drug-target”
heterogeneous network. NRWRH was compared
with traditional supervised or semisupervised
methods such as NRWR (Chen et al. 2012), RWRH
(Li & Patra 2010), and RWR (Camoglu et al. 2005;
Kohler et al. 2008), which makes full use of
network-based information to achieve random
walking on the “drug-target” heterogeneous network
and improve the accuracy of predicting DTIs, but the
method still has certain shortcomings, such as the
problem of randomness, which is mainly caused by
the choice of the initial probability (Ganegoda et al.
2015). LPMIHN (Yan et al. 2016) is a label
propagation method optimized by Yan et al. based
on the NRWRH method. Its “drug-target”
heterogeneous network consists of a drug similarity
network, a target similarity network, and a drug-
target interaction network. Compared with NRWRH,
LPMIHN used a label propagation algorithm on the
constructed “drug-target” heterogeneous network to
infer potential DTIs, which reduces the network
sparsity problem caused by rare drug-target
interactions and further improves the prediction
accuracy. DTINet (Luo et al. 2017) is a
computational prediction pipeline developed by Luo
et al. that integrates multiple drug-related
information. Particularly, this method integrated six
different networks (including a drug-protein
interaction network, a protein similarity network, a
protein-disease association network, a drug-disease
association network, a drug similarity network, and a
drug-side effect association network) into the “drug-
disease-target-side effect” heterogeneous networks
and utilized the restart random walk algorithm to
accurately explain the topological characteristics of
each node in this heterogeneous network. In
addition, in experiments, Luo et al. verified the
novel interaction relationship between the three
drugs and the cyclooxygenase protein predicted by
DTINet and proved the new potential application of
these cyclooxygenase inhibitors in the prevention of
inflammatory diseases. Compared with HNM (Wang
et al. 2014), BLMNII (Mei et al. 2013), NetLapRLS
(Xia Z. et al. 2010), CMF (Xia L. Y. et al. 2019),
DTINet had a better predictive effect, which was
6.9% and 5.9% higher. Shahreza et al. developed a
semisupervised machine learning approach, Heter-
LP, using a label propagation algorithm on the
“drug-target-disease” heterogeneous network (Lotfi
Shahreza et al. 2019). The network of this approach
consists of a drug-disease association network, a
drug-target interaction network, and a disease-target
association network. In particular, Shahreza et al.
applied Heter-LP to analyze innovative putative
drug-disease, drug-target, and disease-target
relationships, including cosyntropin (drug) and
DHCR7, IGF1R, MC1R, MAP3K3, and TOP2A
(protein targets), for a rare disease adrenocortical
carcinoma (ACC). Heter-LP provided a new way for
the treatment of ACC (Lotfi Shahreza et al. 2017).
DHLP-1 (Maleki et al. 2020) and DHLP-2 (Maleki
et al. 2020), with two distributed label propagation
methods based on the “drug-target-disease”
heterogeneous network developed by Maleki et al.
Its heterogeneous network consists of a drug-disease
association network, a drug-target interaction
network, and a disease-target association network.
Compared with the two nondistributed methods,
MINProp (Lotfi Shahreza et al. 2017) and Heter-LP,
the two methods had superior results in terms of
running time and accuracy.
Network-based inference (NBI) is another
common approach to analyze heterogeneous
networks and it is frequently used in the prediction
methods for DTIs. HGBI (Wang et al. 2013) is a
new heterogeneous network-based inference method
proposed by Wang et al. This method constructs a
“drug-target” heterogeneous network by the known
drug-target interaction network, a drug similarity
network, and a target similarity network, and it
predicts DTIs based on this heterogeneous network.
Its prediction accuracy was improved compared with
NBI (Cheng et al. 2012) and BLM (Bleakley &
Yamanishi 2009). Wang et al. developed TL_HGBI
(Wang et al. 2014), which adopts the guilt-by-
association principle to integrate five networks
(including a disease similarity network, a drug-
disease association network, a drug similarity
network, a drug-target interaction network, and a
target similarity network) into the “drug-target-
disease” heterogeneous network. It optimized the
Networks and Algorithms in Heterogeneous Network-based Methods for Drug-target Interaction Prediction: A Survey and Comparison
69

HGBI method, particularly compared with other
methods. When the heterogeneous network model
was changed or the iterative algorithm was updated,
TL_HGBI could not only automatically construct a
new drug-target relationship network but also
automatically add drug-target information for drug-
disease relationship prediction. DT hybrid (domain
tuned-hybrid) (Alaimo et al. 2013) is an NBI
recommendation method based on heterogeneous
networks developed by Alaimo et al., integrating
NBI and Hybrid (Alaimo et al. 2013) tools. The
“drug-target” heterogeneous network of this method
includes a drug similarity network, a target
similarity network, and a drug-target interaction
network. Different from the traditional NBI
recommendation method, the DT hybrid takes into
account the important characteristics of the drug
target domain (Alaimo et al. 2015). SDTNBI (Wu et
al. 2017) is an NBI method based on a “drug-
substructure-target” heterogeneous network
developed by Wu et al. The heterogeneous network
consists of a new chemical entity-substructure
network, a substructure-drug network, and a drug-
target interaction network. This method prioritizes
potential targets of old drugs, failed drugs, and new
chemical entities and combines network and
chemical information to establish relationships
between new chemical entities and known DTI
networks. The advantage of SDTNBI is that it can
predict potential targets of new chemical entities,
whereas traditional network-based methods cannot.
The matrix factorization method can solve the
data sparsity problem well with better prediction
accuracy and has been widely used in the prediction
of DTIs. Liu et al. built a “drug-target”
heterogeneous network by integrating a drug
similarity network, a target similarity network, and a
drug-target interaction network while using the
matrix factorization method to develop NRLMF
(Liu et al. 2016). This method used the
neighborhood regularization logistic matrix
factorization algorithm to establish the interaction
probability model between the drug and the target, in
which the attributes of the drug and the target were
represented by the drug-specific and target-specific
potential vectors, respectively. The average AUC
and AUPR values of NRLMF in the gold standard
dataset are better than those of NetLapRLS (Xia Z.
et al. 2010), BLM-NII (Mei et al. 2013), WNN-GIP
(van Laarhoven & Marchiori 2013), KBMF2K
(Gonen 2012), CMF (Xia L. Y. et al. 2019). KMDR
(Kuang Q. F. et al. 2017) is a heterogeneous network
method based on the kernel matrix reduction
dimension algorithm developed by Kuang et al. The
“drug-target” heterogeneous
Table 1: Drug-target interaction predictions methods.
Name Networks
Algorithm
classification
Algorithms
Datasets for network
construction
Ref
NRWRH
drug-target (protein-protein +
drug-drug + drug-target)
Network
propagation
Random walk with
restarts (RWR)
DrugBank, KEGG,
SuperTarget,
Yamanishi et al.
(Yamanishi et al. 2008)
(Chen et
al. 2012)
HGBI
drug-target (drug-drug + target-
target + drug-target)
Network
inference
Network inference
Sophic
Integrated Druggable
Genome Database
(Sophic 2012), OMIM,
DrugBank, InterPro
(Hunter et al. 2009)
(Wang et
al. 2013)
TL_HGBI
drug-target-disease (disease-
disease + disease-drug + drug-
drug + drug-target + target-
target)
Network
inference
Triple layer
heterogeneous
graph based
inference
DrugBank, Sophic
Integrated Druggable
Genome Database
(Sophic 2012), OMIM,
Gottlieb et al. (Gottlieb
et al. 2011)
(Wang et
al. 2014)
DT-Hybrid
drug-target (drug-drug + target-
target + drug-target)
Network
inference
Bipartite network
projection
DrugBank,
Yamanishi et al.
(Yamanishi et al. 2008)
(Alaimo
et al.
2013)
DASPfind
drug-target (drug-drug + target-
target + drug-target)
Network path
analysis
Simple paths
finding
DrugBank, KEGG,
SuperTarget, BRENDA,
Yamanishi et al.
(Yamanishi et al. 2008)
(Ba-Alawi
et al.
2016)
NRLMF
drug-target (drug-drug + target-
target + drug-target)
Matrix
factorization
Neighborhood
regularized logistic
matrix factorization
Matador, ChEMBL,
DrugBank, KEGG,
SuperTarget, BRENDA
(Liu et al.
2016)
LPMIHN
drug-target (drug-drug + target-
target + drug-target)
Network
propagation
Label propagation
ChEMBL, DrugBank,
KEGG, SuperTarget,
(Yan et al.
2016)
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
70

BRENDA
KMDR
drug-target (drug-drug + target-
target + drug-target)
Matrix
factorization
Kernel matrix
dimension
reduction
DrugBank, KEGG,
UniProt
(Kuang Q.
F. et al.
2017)
DTINet
drug-disease-target-side effect
(drug-protein + protein-protein
+ protein-disease + disease-
drug + drug-drug + drug-side
effect)
Network
propagation
RWR and diffusion
component analysis
DrugBank, HPRD, CTD,
SIDER
(Luo et al.
2017)
DNILMF
drug-target (drug-drug + target-
target + drug-target)
Matrix
factorization
A dual-network
integrated logistic
matrix factorization
DrugBank, KEGG,
BRENDA, SuperTarget,
COMPOUND
(Hao et al.
2017)
GRMF
drug-target (drug-drug + target-
target + drug-target)
Matrix
factorization
Graph regularized
matrix factorization
Yamanishi et al.
(Yamanishi et al. 2008)
(Ezzat et
al. 2017)
SDTNBI
substructure-drug-target
(drug-substructure + drug-
target +
new chemical entity-
substructure)
Network
inference
Network inference
ChEMBL, DrugBank,
BindingDB
(Wu et al.
2017)
Heter-LP
drug-target-disease (drug-
disease +
drug-target + disease-target)
Network
propagation
Label propagation DrugBank, SuperTarget
(Lotfi
Shahreza
et al.
2019)
DHLP-1
DHLP-2
drug-target-disease (drug-
disease +
drug-target + disease-target)
Network
propagation
Label propagation
Yamanishi et al.
(Yamanishi et al. 2008)
(Maleki et
al. 2020)
iDrug
drug-target-disease (drug-target
+ drug-disease + drug-drug +
targe
t
-target + disease-disease)
Matrix
factorization
Matrix factorization
CTD, Gottlieb et al.
(Gottlieb et al. 2011)
(Chen H.
et al.
2020)
network of KMDR consists of a drug similarity
network, a target similarity network, and a drug-
target interaction network. KMDR can reduce the
prediction bias, and it has a better DTI performance
than the regularized least squares classifier (RLS)
(Kuang Q. et al. 2014) and a semisupervised link
prediction classifier (SLP) (Kuang Q. et al. 2014).
DNILMF (Hao et al. 2017) is a dual-network
integrated logistic matrix factorization algorithm
developed by Hao et al., and its “drug-target”
heterogeneous network consists of a drug similarity
network, a target similarity network, and a drug-
target interaction network. This method used a
domain regularization logistic matrix factorization
algorithm, which was optimized based on NRLMF,
to improve the drug-target prediction accuracy, and
its prediction results had higher AUC and AUPR
values than NRLMF. Ezzat et al. developed a
network-based regularized matrix decomposition
tool, GRMF (Ezzat et al. 2017), whose “drug-target”
heterogeneity network consists of a drug similarity
network, a target similarity network, and a drug-
target interaction network.In addition, this method
took into account the situation in which many non-
occurring edges in the network were unknown or
missing cases and it added edges with intermediate
interaction probability scores in the preprocessing
step to improve the prediction results of the new
drugs and new targets. As a result, GRMF
performed very well in predicting the left-out
interactions. Chen et al. integrated a drug-target
interaction network, a drug-disease association
network, a drug similarity network, a disease
similarity network, and a target similarity network to
form the “drug-disease-target” heterogeneous
network and thus developed the iDrug (Chen H. et
al. 2020) method. This method utilized a matrix
factorization method to connect a drug-disease
association network and a drug-target interaction
network through drugs. MBiRW has better drug-
target prediction and drug-disease prediction
performance than TH_HGBI, and it can also identify
new drug-miRNA interactions.
In addition to the above three types of DTI
prediction methods, there are other methods, such as
network path analysis. DASPfind (Ba-Alawi et al.
2016) is a network path analysis method based on a
heterogeneous network developed by Ba-Alawi et al.
Its “drug-target” heterogeneous network consists of
a drug similarity network, a target similarity
network, and a drug-target interaction network.
Compared with the other methods, the advantage of
this method is that it can better predict DTIs with
unknown targets or drugs with fewer targets and it
has a better prediction performance than HGBI, DT-
Hybrid, and NRWRH.
Networks and Algorithms in Heterogeneous Network-based Methods for Drug-target Interaction Prediction: A Survey and Comparison
71

3 RESULTS AND DISCUSSION
3.1 Comparison of DTIs Prediction
Methods
Common algorithm evaluation methods include
independent dataset testing, ab initio prediction,
leave-one-out verification, and external dataset
verification, and the most commonly used cross-
validation is the tenfold cross-validation method,
which is widely used in the evaluation of algorithm
accuracy. The AUC value is the area under the ROC
curve, which can usually indicate the overall
performance of the algorithm, and it can be used to
compare the relative performance of different
algorithms, with larger values indicating better
algorithm performance (Sing et al. 2005). The PR
curve (precision recall curve) shows the relationship
between precision and recall. In most of the
literature, the indicators used to evaluate the
prediction performance of the algorithm are the area
under the curve (AUC) and the area under the
precision recall curve (AUPR) (Nascimento et al.
2016).
We collected the AUC and AUPR values of
more than a dozen methods, including NRWRH,
DT-Hybrid, DHLP, etc. on the gold standard dataset
(Lotfi Shahreza et al. 2018) (http://web.kuicr.kyoto-
u.ac.jp/supp/yoshi/drugtarget/), which was divided
into four parts (Yamanishi et al. 2008): enzyme, ion
channel, GPCR, and nuclear receptor (Table 2).
As shown in Table 2, DT-Hybrid, LPMIHN,
DNILMF, MINProp, NRLMF, and SDTNBI have
high AUC values on the same benchmark datasets,
and their AUC values on the four parts of
benchmark datasets are above 90%. In particular,
DT-Hybrid has a high prediction accuracy of DTIs
with AUC values of approximately 99% on the four
types of benchmark datasets. In addition, LPMIHN
has the highest AUPR value and has better
application prospects.
3.2 Comparison of Heterogeneous
Networks and Algorithms
In addition to the above comparison of the
performance of the DTI prediction methods through
the AUC and AUPR values, this paper also provided
statistics and comparisons of the effects of different
heterogeneous networks and different algorithms on
the prediction results (Table 3).
The comparisons shown in Table 3 indicate that
most of the methods with higher accuracy in
predicting DTIs used “drug-target” heterogeneous
networks constructed by a drug similarity network, a
target similarity network, and a drug-target
interaction network. The “drug-disease-target”
heterogeneous network constructed by adding
disease information did not contribute significantly
to an improvement of the prediction accuracy. Using
the same “drug-target” heterogeneous network, the
prediction accuracy of DNILMF and NRLMF using
the logistic matrix-based decomposition method was
higher than that of GRMF using only the matrix
decomposition method, and the AUPR value
increased from 76.3% to more than 98%. Therefore,
logistic matrix factorization was chosen as superior
for the prediction method of DTIs based on
heterogeneous networks. In addition, among the
methods using “drug-target” heterogeneous
networks, network propagation methods and
network inference methods were used for better
prediction.
Table 2: Reported AUC and AUPR on gold standard datasets in literature.
Method
Enzyme
Ion channel GPCR Nuclear receptor
Ref
AUC AUPR AUC AUPR AUC AUPR AUC AUPR
NRWRH 0.953 - 0.971 - 0.945 - 0.867 - (Chen et al. 2012)
DT-
Hybrid
0.999
- 0.997 -
0.999
-
1.000
-
(Eslami
Manoochehri &
Nourani 2020)
SDTNBI 0.958 - 0.971 - 0.966 - 0.932 - (Wu et al. 2017)
DASPfin
d
0.929 - 0.907 - 0.881 - 0.853 -
(Eslami
Manoochehri &
Nourani 2020)
HGBI 0.916 - 0.889 - 0.913 - 0.876 -
(Eslami
Manoochehri &
Nourani 2020)
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
72

NRLMF 0.987 0.892 0.989 0.906 0.969 0.749 0.950 0.728 (Liu et al. 2016)
LPMIHN
0.999
0.929
0.998
0.961
0.998
6
0.973
0.996
0.970
(Yan et al. 2016)
GRMF - 0.763 - 0.745 - 0.567 - 0.423 (Ezzat et al. 2017)
DHLP-1 - - - - 0.976 0.766 - - (Maleki et al. 2020)
DLHP-2 - - - - 0.955 0.956 - - (Maleki et al. 2020)
HeterLP - - - - 0.967 0.796 - - (Maleki et al. 2020)
DNILMF 0.989 0.922 0.990 0.938 0.975 0.821 0.955 0.751 (Hao et al. 2017)
4 CONCLUSIONS
In this paper, we systematically reviewed the
heterogeneous network-based prediction methods
for DTIs, and the statistical analysis of the
heterogeneous networks showed that most of the
DTI prediction methods used the “drug-target”
heterogeneous network, which was comprised of a
drug similarity network, a target similarity network,
and a drug-target interaction network. In terms of the
algorithm selection methods, network inference,
network propagation and matrix factorization were
used for the prediction of DTIs. By comparing the
performance of these DTI methods against the gold
standard dataset, DT-Hybrid and LPMIHN were
found to have the best prediction performance. By
comparing the heterogeneous networks and
algorithms against the gold standard dataset, it was
found that the method using “drug-target”
heterogeneous networks had better prediction
performance and that the triple-layer heterogeneous
networks constructed by adding disease information
were of limited use in improving the prediction
accuracy. Among the “drug-target” heterogeneous
networks, network propagation and network
inference methods were found to have better
prediction performance.
ACKNOWLEDGEMENTS
This work was funded by the Natural Science
Foundation of Chongqing (cstc2019jcyj-
msxmX0271); the Science and Technology Research
Program of Chongqing Municipal Education
Commission (KJQN202100642).
REFERENCES
Abbou, S. I., Bouziane, H., & Chouarfia, A. (2021).
Logistic matrix factorisation and generative
adversarial neural network-based method for
predicting drug-target interactions. Mol Divers, 25 (3),
1497-1516.
Alaimo, S., Bonnici, V., Cancemi, D., et al. (2015). DT-
Web: a web-based application for drug-target
interaction and drug combination prediction through
domain-tuned network-based inference. BMC Syst
Biol, 9 Suppl 3, S4.
Alaimo, S., Pulvirenti, A., Giugno, R., et al. (2013). Drug-
target interaction prediction through domain-tuned
network-based inference. Bioinformatics, 29 (16),
2004-2008.
Ashburn, T. T., & Thor, K. B. (2004). Drug repositioning:
identifying and developing new uses for existing
drugs. Nature Reviews Drug Discovery, 3 (8), 673-
683.
Ba-Alawi, W., Soufan, O., Essack, M., et al. (2016).
DASPfind: new efficient method to predict drug-target
interactions. Journal of Cheminformatics, 8, 15.
Bi, C., Beeram, S., Li, Z., et al. (2015). Kinetic analysis of
drug-protein interactions by affinity chromatography.
Drug Discov Today Technol, 17, 16-21.
Bleakley, K., & Yamanishi, Y. (2009). Supervised
prediction of drug-target interactions using bipartite
local models. Bioinformatics, 25 (18), 2397-2403.
Camoglu, O., Can, T., Singh, A. K., et al. (2005). Decision
tree based information integration for automated
protein classification. Journal of Bioinformatics and
Computational Biology, 3 (3), 717-742.
Chen, H., Cheng, F., & Li, J. (2020). iDrug: Integration of
drug repositioning and drug-target prediction via
cross-network embedding. PLoS Computational
Biology, 16 (7), e1008040.
Chen, X., Liu, M. X., & Yan, G. Y. (2012). Drug-target
interaction prediction by random walk on the
heterogeneous network. Molecular Biosystems, 8 (7),
1970-1978.
Chen, Z. H., You, Z. H., Guo, Z. H., et al. (2020).
Prediction of Drug-Target Interactions From Multi-
Molecular Network Based on Deep Walk Embedding
Model. Front Bioeng Biotechnol, 8, 338.
Networks and Algorithms in Heterogeneous Network-based Methods for Drug-target Interaction Prediction: A Survey and Comparison
73

Cheng, F., Liu, C., Jiang, J., et al. (2012). Prediction of
drug-target interactions and drug repositioning via
network-based inference. PLoS Computational
Biology, 8 (5), e1002503.
Engin, H. B., Gursoy, A., Nussinov, R., et al. (2014).
Network-based strategies can help mono- and poly-
pharmacology drug discovery: a systems biology
view. Current Pharmaceutical Design, 20 (8), 1201-
1207.
Eslami Manoochehri, H., & Nourani, M. (2020). Drug-
target interaction prediction using semi-bipartite graph
model and deep learning. BMC Bioinformatics, 21
(Suppl 4), 248.
Ezzat, A., Wu, M., Li, X. L., et al. (2019). Computational
prediction of drug-target interactions using
chemogenomic approaches: an empirical survey. Brief
Bioinform, 20 (4), 1337-1357.
Ezzat, A., Zhao, P., Wu, M., et al. (2017). Drug-Target
Interaction Prediction with Graph Regularized Matrix
Factorization. IEEE/ACM Transactions on
Computational Biology Bioinformatics, 14 (3), 646-
656.
Ganegoda, G. U., Li, M., Wang, W., et al. (2015).
Heterogeneous network model to infer human disease-
long intergenic non-coding RNA associations. IEEE
Transactions on Nanobioscience, 14 (2), 175-183.
Gonen, M. (2012). Predicting drug-target interactions
from chemical and genomic kernels using Bayesian
matrix factorization. Bioinformatics, 28 (18), 2304-
2310.
Gottlieb, A., Stein, G. Y., Ruppin, E., et al. (2011).
PREDICT: a method for inferring novel drug
indications with application to personalized medicine.
Molecular Systems Biology, 7, 496.
Hao, M., Bryant, S. H., & Wang, Y. (2017). Predicting
drug-target interactions by dual-network integrated
logistic matrix factorization. Scientific Reports, 7,
40376.
Hodos, R. A., Kidd, B. A., Shameer, K., et al. (2016). In
silico methods for drug repurposing and
pharmacology. Wiley Interdisciplinary Reviews-
Systems Biology and Medicine, 8 (3), 186-210.
Hunter, S., Apweiler, R., Attwood, T. K., et al. (2009).
InterPro: the integrative protein signature database.
Nucleic Acids Research, 37 (Database issue), D211-
215.
Jacob, L., & Vert, J. P. (2008). Protein-ligand interaction
prediction: an improved chemogenomics approach.
Bioinformatics, 24 (19), 2149-2156.
Kim, S., Chen, J., Cheng, T., et al. (2021). PubChem in
2021: new data content and improved web interfaces.
Nucleic Acids Research, 49 (D1), D1388-D1395.
Kohler, S., Bauer, S., Horn, D., et al. (2008). Walking the
interactome for prioritization of candidate disease
genes. American Journal of Human Genetics, 82 (4),
949-958.
Kuang, Q., Wang, M., Li, R., et al. (2014). A systematic
investigation of computation models for predicting
Adverse Drug Reactions (ADRs). PLoS One, 9 (9),
e105889.
Kuang, Q. F., Li, Y. Z., Wu, Y. M., et al. (2017). A kernel
matrix dimension reduction method for predicting
drug-target interaction. Chemometrics and Intelligent
Laboratory Systems, 162, 104-110.
Li, Y., & Patra, J. C. (2010). Genome-wide inferring gene-
phenotype relationship by walking on the
heterogeneous network. Bioinformatics, 26 (9), 1219-
1224.
Liu, Y., Wu, M., Miao, C., et al. (2016). Neighborhood
Regularized Logistic Matrix Factorization for Drug-
Target Interaction Prediction. PLoS Computational
Biology, 12 (2), e1004760.
Lotfi Shahreza, M., Ghadiri, N., & Green, J. R. (2019).
Heter-LP: A Heterogeneous Label Propagation
Method for Drug Repositioning. Methods in
Molecular Biology, 1903, 291-316.
Lotfi Shahreza, M., Ghadiri, N., Mousavi, S. R., et al.
(2017). Heter-LP: A heterogeneous label propagation
algorithm and its application in drug repositioning.
Journal of Biomedical Informatics, 68, 167-183.
Lotfi Shahreza, M., Ghadiri, N., Mousavi, S. R., et al.
(2018). A review of network-based approaches to drug
repositioning. Brief Bioinform, 19 (5), 878-892.
Luo, Y., Zhao, X., Zhou, J., et al. (2017). A network
integration approach for drug-target interaction
prediction and computational drug repositioning from
heterogeneous information. Nature Communications,
8 (1), 573.
Maleki, E. F., Ghadiri, N., Shahreza, M. L., et al. (2020).
DHLP 1&2: Giraph based distributed label
propagation algorithms on heterogeneous drug-related
networks. Expert Systems with Applications, 159.
Mei, J. P., Kwoh, C. K., Yang, P., et al. (2013). Drug-
target interaction prediction by learning from local
information and neighbors. Bioinformatics, 29 (2),
238-245.
Nagamine, N., Shirakawa, T., Minato, Y., et al. (2009).
Integrating statistical predictions and experimental
verifications for enhancing protein-chemical
interaction predictions in virtual screening. PLoS
Computational Biology, 5 (6), e1000397.
Nascimento, A. C., Prudencio, R. B., & Costa, I. G.
(2016). A multiple kernel learning algorithm for drug-
target interaction prediction. BMC Bioinformatics, 17,
46.
Paul, S. M., Mytelka, D. S., Dunwiddie, C. T., et al.
(2010). How to improve R&D productivity: the
pharmaceutical industry's grand challenge. Nature
Reviews Drug Discovery, 9 (3), 203-214.
Pliakos, K., & Vens, C. (2020). Drug-target interaction
prediction with tree-ensemble learning and output
space reconstruction. BMC Bioinformatics, 21 (1), 49.
Sachdev, K., & Gupta, M. K. (2019). A comprehensive
review of feature based methods for drug target
interaction prediction. Journal of Biomedical
Informatics, 93, 103159.
Saint-Antoine, M. M., & Singh, A. (2020). Network
inference in systems biology: recent developments,
challenges, and applications. Current Opinion
Biotechnology, 63, 89-98.
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
74

Sing, T., Sander, O., Beerenwinkel, N., et al. (2005).
ROCR: visualizing classifier performance in R.
Bioinformatics, 21 (20), 3940-3941.
Sophic. (2012).
http://www.sophicalliance.com/documents/sophicdocs
/White%20Paper%20Update%201-27-
11/The%20Druggable%20Genome012511.pdf.
van Laarhoven, T., & Marchiori, E. (2013). Predicting
Drug-Target Interactions for New Drug Compounds
Using a Weighted Nearest Neighbor Profile. PLoS
One, 8 (6), e66952.
Wang, W., Yang, S., & Li, J. (2013). Drug target
predictions based on heterogeneous graph inference.
Pacific Symposium on Biocomputing, 53-64.
Wang, W., Yang, S., Zhang, X., et al. (2014). Drug
repositioning by integrating target information through
a heterogeneous network model. Bioinformatics, 30
(20), 2923-2930.
Wu, Z., Cheng, F., Li, J., et al. (2017). SDTNBI: an
integrated network and chemoinformatics tool for
systematic prediction of drug-target interactions and
drug repositioning. Briefings in Bioinformatics, 18 (2),
333-347.
Wu, Z., Li, W., Liu, G., et al. (2018). Network-Based
Methods for Prediction of Drug-Target Interactions.
Front Pharmacol, 9, 1134.
Xia, L. Y., Yang, Z. Y., Zhang, H., et al. (2019). Improved
Prediction of Drug-Target Interactions Using Self-
Paced Learning with Collaborative Matrix
Factorization. Journal of Chemical Information and
Modeling, 59 (7), 3340-3351.
Xia, Z., Wu, L. Y., Zhou, X., et al. (2010). Semi-
supervised drug-protein interaction prediction from
heterogeneous biological spaces. BMC Systems
Biology, 4 Suppl 2, S6.
Yamanishi, Y., Araki, M., Gutteridge, A., et al. (2008).
Prediction of drug-target interaction networks from the
integration of chemical and genomic spaces.
Bioinformatics, 24 (13), i232-240.
Yan, X. Y., Zhang, S. W., & Zhang, S. Y. (2016).
Prediction of drug-target interaction by label
propagation with mutual interaction information
derived from heterogeneous network. Mol Biosyst, 12
(2), 520-531.
Zhao, Q., Yu, H., Ji, M., et al. (2019). Computational
Model Development of Drug-Target Interaction
Prediction: A Review. Curr Protein Pept Sci, 20 (6),
492-494.
Networks and Algorithms in Heterogeneous Network-based Methods for Drug-target Interaction Prediction: A Survey and Comparison
75
