
Semantic-based Data Integration and Mapping Maintenance:
Application to Drugs Domain
Mouncef Naji
1
, Maroua Masmoudi
2
, Hajer Baazaoui Zghal
1
, Chirine Ghedira Guegan
3
,
Vlado Stankovski
4
and Dan Vodislav
1
1
ETIS Labs, CY Cergy Paris University, ENSEA / CNRS, France
2
CY Tech, Pau, CY Cergy Paris University, France
3
Univ. Lyon, Universit
´
e Jean-Moulin Lyon 3, LIRIS UMR5205, iaelyon School of Management, France
4
Faculty of Civil and Geodetic Engineering, University of Ljubljana, Ljubljana, Slovenia
Keywords:
Big Data, Mapping Maintenance, Data Integration, Ontology, Deep Learning, Classification.
Abstract:
In recent years, the number of data sources and the amount of generated data are increasing continuously. This
voluminous data leads to several issues of storage capacities, data inconsistency, and difficulty of analysis.
In the midst of all these difficulties, data integration techniques try to offer solutions to optimally face these
problems. In addition, adding semantics to data integration solutions has proven its utility for tackling these
difficulties, since it ensures semantic interoperability. In our work, which is placed in this context, we propose
a semantic-based data integration and mapping maintenance approach with application to drugs domain. The
contributions of our proposal deal with 1) a virtual semantic data integration and 2) an automated mapping
maintenance based on deep learning techniques. The goal is to support the continuous and occasional data
sources changes, which would highly affect the data integration. To this end, we focused mainly on managing
metadata change within an integrated structure, refereed to as mapping maintenance. Our deep learning models
encapsulate both convolutional, and Long short-term memory networks. A prototype has been developed and
performed on two use cases. The process is fully automated and the experiments show significant results
compared to the state of the art.
1 INTRODUCTION
Big data has become a great phenomenon in the mod-
ern industry, especially with the availability of enor-
mous amounts of data. The benefits of such data
sparkled the curiosity of multiple companies and re-
searchers.
Nevertheless, although it offers such benefits,
managing the different data sources remains a chal-
lenging problem. Indeed, data size, heterogeneity, va-
riety, etc, make it hard to manage and interlink data
sources. Accordingly, to overcome this problem, it
has been suggested to add semantics to data. Ontolo-
gies, as a knowledge representation formalism, con-
tinue to be a promising solution for the semantic in-
teroperability between this data. Indeed, they allow
representing data following a semantic model, which
permit a better understanding of data. In this regard,
in terms of heterogeneity, variety and volume, drugs
data are one of the targeted fields by big data tech-
niques. In effect, big biomedical data has grown expo-
nentially during the last decades and a similar growth
rate is expected in the next years, resulting into differ-
ent interoperability conflicts among data collections
with entities being dispersed across different datasets.
Each data source can likely get a change of structure
and metadata, which compromises the whole integra-
tion (Maria-Esther and al., 2019). As aforesaid, the
data sources being numerous and varied, this issue
impacts consistency in virtual data integration and its
maintenance. This latter, namely, mapping mainte-
nance is defined as the task aiming to keep existing
mappings in an updated and valid state, reflecting the
changes affecting a source’s entities at evolution time
(Cesar and al., 2015).
Therefore, this work aims to integrate data sources
based on ontologies with an application to drugs’ do-
main. Our proposal takes into account both the data
sources integration changes and associated metadata.
To do so, the study focuses on automatically manip-
ulating mappings generated from virtual data integra-
tion, in order to adapt them to the observed changes.
Thus, the study responds to two main research ques-
tions: (i) which data integration is suitable for the
Naji, M., Masmoudi, M., Zghal, H., Guegan, C., Stankovski, V. and Vodislav, D.
Semantic-based Data Integration and Mapping Maintenance: Application to Drugs Domain.
DOI: 10.5220/0011279900003266
In Proceedings of the 17th International Conference on Software Technologies (ICSOFT 2022), pages 469-477
ISBN: 978-989-758-588-3; ISSN: 2184-2833
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
469

mentioned problem, and (ii) how to adapt and update
mappings to handle data sources’ changes. The orig-
inality of our work is the novel automated mapping
maintenance that we propose, based on deep learning
techniques. This article is organized as follows: Sec-
tion 2 presents related works. Our proposal is detailed
in section 3. Section 4 presents the implementation
and discusses the evaluation of our proposal. Lastly,
we conclude and deliver the future works in section 5.
2 STATE OF THE ART
Our study of recent works has focused on schema
matching and mapping maintenance as a solution for
change in the metadata of data sources.
Schema matching makes use of name, descrip-
tion, type of data, constraint, and schema struc-
ture in order to identify the match between two at-
tributes of data source’s schema. (Do, 2007) de-
scribes the architecture, functionality, and evaluation
of the schema matching system COMA++ (Combin-
ing Matchers). COMA++ represents a generic and
customised system for semi-automatic schema match-
ing, which combines different match algorithms in a
flexible way. But even though the work shows good
results, it stays limited due to the unavailability of
metadata and schema of all data-sources. Further-
more, the use of metadata is not always appropriate to
schema matching process. This issue has been related
in many works. That being said, work on schema
matching has shifted towards Instance based match-
ing. This approach employs the available instances
as a source to identify the correspondences between
schema attributes. (Munir and al, 2014) focuses
on finding similarities/matchings between databases
based on the instance approach. It uses the databases’
primary keys. The proposed approach consists of two
main phases : Row Similarity and Attribute Similar-
ity. This work shows significant results in terms of
matching accuracy. However, it remains limited by
its applicability to only databases.
While working on databases, (Sahay and al, 2019)
and (Mehdi and al, 2015) use a hybrid approach, using
schemas and data provided to manage matchings, by
using machine learning. They try to work on one-
to-one matchings, and introduce a dictionary for one-
to-many matchings. The results seem sufficient for
simple use cases, but show mediocre performance in
a complex mappings while supporting only databases.
Concerning other formats, (Mohamed-Amine and
al, 2017) work on Json datasets. They focus solely
on schema inference, but they show great potential
for schema matching. They deal with the problem
of inferring a schema from massive JSON datasets,
by identifying a JSON type language which is simple
and expressive enough to capture irregularities and
to give complete structural information about input
data. (Ahmad Abdullah Alqarni, 2017) introduces
two approaches to improve XML Schema matching
efficiency: internal filtering and node inclusion. In
the former, it detects dissimilar elements at all schema
levels at early stages of the matching process. And In
the latter, it detects dissimilarity between elements on
leaf nodes only using their ancestor degree of simi-
larity and then exclude them from the next phase of
matching process.
In another context, several researches focused on
mappings maintenance to fix data availability. This
means manipulating and updating mappings to handle
data sources’ and ontologies’ change.
In that matter, (Cesar and al, 2015) and
(N. Popitsch, 2018) take into account the definition
of established mappings, the evolution of Knowl-
edge Organization Systems (KOS) and the possible
changes that can be applied to the mappings. they de-
fine complete frameworks based on formal heuristics
that drives the adaptation of KOS mappings. These
studies show great results in terms of mappings main-
tenance, but they require a lot of human interven-
tion, with a repeated the integration process, which
re-value the need for a mapping maintenance solution.
(Khattaka and al., 2015) proposes an other ap-
proach towards mappings evolution regeneration. It
focuses on finding alignments between ontologies.
The paper proposes a mapping reconciliation ap-
proach between the updated ontologies that have been
found to take less time to process compared to the
time of existing systems when only the changed re-
sources are considered and also eliminates the stale-
ness of the existing mappings. Even though this work
presents intresting work concerning mappings main-
tenance, it focuses only on ontology descriptions,
without taking into consideration the data change.
2.0.1 Synthesis
The literature revue on schema matching and map-
ping maintenance shows that the existing works fo-
cused either on manipulating ontologies or by chang-
ing data sets. But since our focus is on mapping files,
no study has tried to manipulate the files directly. That
being said, our aim is to focus on how to manipulate
and update mapping files, following a change in data
sources’ metadata.
ICSOFT 2022 - 17th International Conference on Software Technologies
470
3 SEMANTIC-BASED DATA
INTEGRATION AND MAPPING
MAINTENANCE APPROACH
In this section, we describe our proposed approach
for drug integration. The aim is to provide a virtual
semantic data integration of heterogeneous multiple
data sources in drugs domain. As aforementioned, the
proposed approach focuses on two main functionali-
ties: Federated data integration and mappings update.
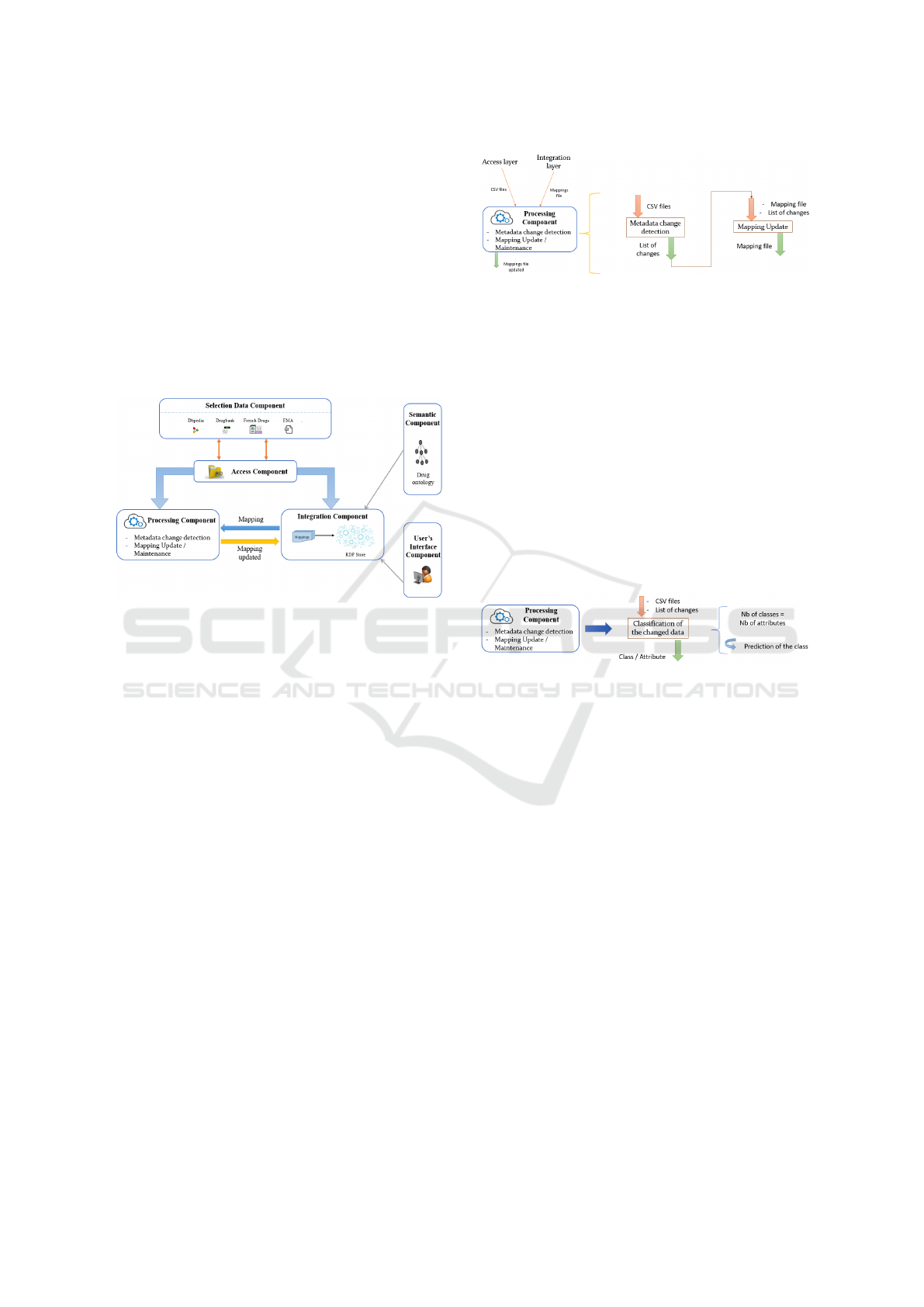
Figure 1 presents the global architecture of the
proposed approach. The layered architecture is com-
posed of 6 components:
Figure 1: The proposed data integration architecture.
• Selection Data Component: It encompasses the
selection of different data sources with different
contents and data types (XML, CSV, TXT, etc).
• Access Component: It contains all the necessary
implementations allowing the access to multi-
source data.
• Integration Component: The purpose of the in-
tegration component is to combine data coming
from multiple data sources to provide a unified,
single view of data. The integration process is
based on a drug ontology (concepts). The out-
put of the integration is a file of mappings. This
component takes as input, the data sources and the
domain ontology. As output, it returns the map-
pings file for the virtual integration. We point out
that a virtual integration approach means that data
remains in local sources and is accessed through
an intermediate infrastructure called a mediator
(Masmoudi and al, 2021).
• Processing Component: The aim of this compo-
nent is to maintain an integration system up-to-
date. It checks changes in data sources, and up-
dates accordingly the mapping file, as presented
in figure 2.
We perform this processing by following three ba-
sic functionalities:
Figure 2: Processing component.
– Metadata Changes Detection: an algorithm
is proposed to detect changes occurred in the
sources’ metadata. The changes are taken into
account by the algorithm, to classify them into
the corresponding metadata: matching between
and new data sources/metadata.
– Mappings Update: for this functionality, we
develop an algorithm to update the previous
mapping file to match the new changes. It takes
as input the list of changes resulted from the
first algorithm, and outputs the updated map-
ping file.
According to the list of changed attributes, we
use a classification algorithm to match data
with the corresponding attribute (figure 3).
Figure 3: Classification.
– Integration Update: After generating the up-
dated mapping file, this latter gets transferred
to the integration component once more, in or-
der to maintain the integration process with the
new changes.
• Semantic Component: This component contains
the domain ontology allowing the semantic inte-
gration.
• User’s Interface Component: It represents the
front-end interface allowing to dialog with the
proposed system. Through this interface, users
are able to query the integrated data.
3.1 Data and Algorithms’ Description
This section describes the algorithms developed for
our approach, as well as a presentation of the drugs
dataset used for the prototype.
Semantic-based Data Integration and Mapping Maintenance: Application to Drugs Domain
471
3.1.1 Drug Dataset Overview
As aforementioned we applied our approach to the
drugs domain, thus we used the DrugBank dataset
1
.
It is a comprehensive online database containing in-
formation on drugs and drug targets. DrugBank com-
bines detailed drug (i.e. chemical, pharmacological
and pharmaceutical) data with comprehensive drug
target (i.e. sequence, structure, and pathway) infor-
mation. The database contains 9591 drug entries in-
cluding 2037 FDA-approved small molecule drugs,
241 FDA-approved biotech (protein/peptide) drugs,
96 nutraceuticals and over 6000 experimental drugs.
It contains 55 attributes, each attribute describing a
characteristic of the drugs. We enumerate some of
them : the drug identifier (Drugbank-id), the drug’s
name, the drug’s Chemical Abstracts Service number
(CAS number).
3.1.2 Data Integration
The proposed approach starts with semantically inte-
grating data and generating the corresponding map-
ping files. Indeed, it matches the attributes of the data
source (metadata) with the ontology concepts. To do
so, we used Onto-Kit (Masmoudi and al, 2021), an
automatic ontology-based data integration tool. By
importing the drug ontology and the dataset to the
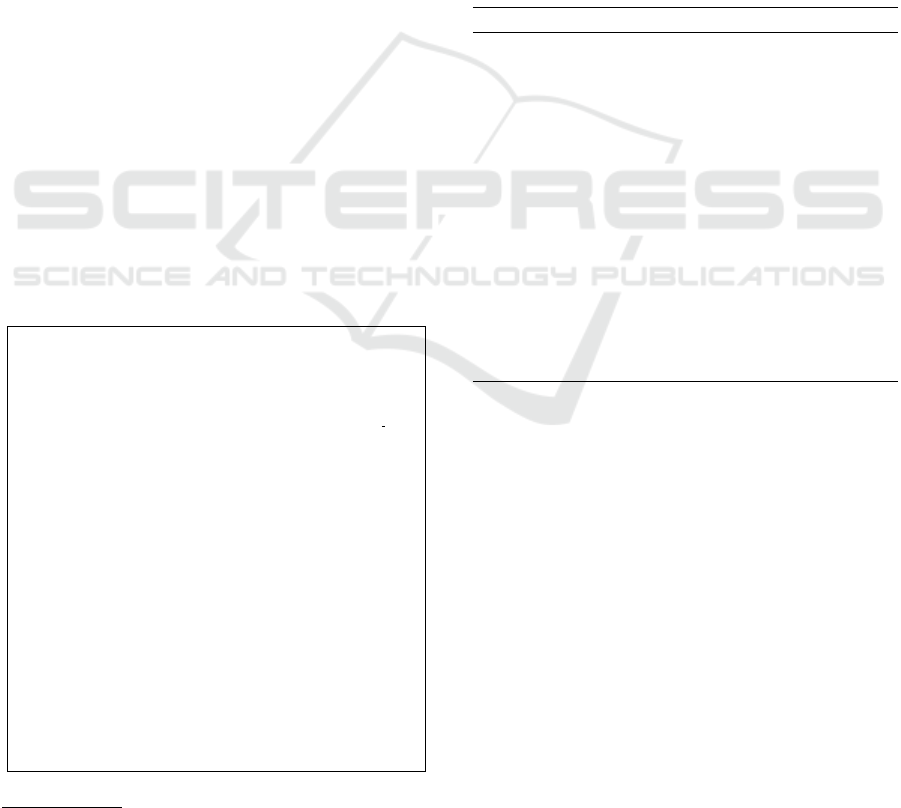
tool, we generate an RML mapping document 1. We
point out that RML is a mapping language that de-
scribes mapping rules between any form of data to
RDF (A triple based graph model language) (Mas-
moudi and al, 2021).
@prefix r r : <h t t p : / / www. w3 . o rg / n s / r 2 r m l #>.
@prefix r m l : <h t t p : / / semweb . mmlab . be / n s / rml#> .
@prefix x s d : <h t t p : / / www. w3 . o r g / 2 0 0 1 / XMLSchema#>.
@prefix r r x : <h t t p : / / www. w3 . o r g / n s / r 2 r m l −e x t #>.
@prefix r r x f : <h t t p : / / www. w3 . o r g / n s / r 2 r m l −e x t / f u n c t i o n s
/ d e f / > .
@prefix d rug b a n k v o c ab : <h t t p : / / b i o 2 r d f . o r g / d r u g b a n k
v o c a b u l a r y : >.
@base <h t t p : / / exa m p le . com / b a s e> .
<#DrugBankMapping> a r r : T ri p l e s M a p ;
rml : l o g i c a l S o u r c e [
rml : s o u r c e ”C: \ U s e r s \marmo\ One D riv e \ B u r eau
\TEST\ d r u g b a n k . c s v ” ;
rml : r e f e r e n c e F o r m u l a t i o n q l : CSV ;
] ;
r r : s u b j e c t M ap [
r r : t e m p l a t e ” h t t p : / / e x ampl e . com / { dr u g bank −i d } ” ;
r r : c l a s s d r u gba n k v o c a b : d r u gban k −i d ;
] ;
r r : p r e d i c a t e O b j e c t M a p [
r r : p r e d i c a t e d r u g b a n k voc a b : name ;
r r : o b j e c t M a p [
rml : r e f e r e n c e ”name ” ; ] ;
] ;
r r : p r e d i c a t e O b j e c t M a p [
r r : p r e d i c a t e d r u g b a n k voc a b : d e s c r i p t i o n ;
r r : o b j e c t M a p [
rml : r e f e r e n c e ” d e s c r i p t i o n ” ; ] ;
] ;
Listing 1: Mapping file.
1
Drugbank dataset, https://go.drugbank.com
This file defines links as a triple map format, be-
tween a concept of the ontology and an attribute of
the data source.
Onto-kit maps each class of the ontology to the
corresponding attribute of the dataset. Then, run-
ning through all the attributes, Onto-kit generates the
predicate-object mapping file, in which the number of
maps corresponds to the number of attributes of the
data source.
3.1.3 Metadata Change Detection
While ensuring the data integration, the work of the
processing component comes in play, when a change
is detected in the data source. We try periodically to
check change in the metadata following the algorithm
1. This detection is based on the old metadata previ-
ously registered of the data source and the new one.
Algorithm 1: Metadata change detection.
1: Old. f ile ← read −CSV (old. f ile)
2: New. f ile ← read −CSV (new. f ile) Read the data
3: List.changes ← []
4: Old.attributes ← metadata − o f (Old. f ile)
5: New.attributes ← metadata.o f (New. f ile) Get the metadata /
attributes
6: for attribute.1 in Old.attributes do
7: for attribute.2 in New.attributes do
8: while attribute.1 and attribute.2 have the same index do
9: if attribut e.1 is not similar to attribute.2 then Check for
changes
10: Add attribut e.2 to List.changes
11: end if
12: end while
13: end for
14: end for
return List.changes
The algorithm 1 compares the old and the new
data source’s metadata, and returns a list of the
changed attributes. The next step is to determine the
corresponding attributes of these changes in the new
data source.
3.1.4 Metadata Matching
With the algorithm 1, we got to detect the attributes
that have been changed in the new data source. But
the problem is that we don’t know to which attributes
correspond these changes.
For example, we know that the metadata
drugbank − id and description have been changed,
but how can we match the new changes to them?
a bad scenario would be to correspond the data ”
DB00105 ” to the attribute description, and the data
” Lepirudin is used to break up clots and to reduce
thrombocytopenia. It binds to thrombin ... ” to
ICSOFT 2022 - 17th International Conference on Software Technologies
472

drugbank − id.
Such a bad matching would have bad impact on
the mapping file, and thus, it would result a wrong
data integration.
Thus, the problem we are facing now, is a classi-
fication one. This means that we want to classify the
data to a corresponding attribute. In other words, we
consider each attribute of the dataset as a class, and
based on the data, we predict the corresponding class.
Therefor, the purpose of the algorithm 2 is to
match the old changed attributes with the new ones.
This is done by training learning models to predict
which class an attribute’s data corresponds to. The
algorithm returns the attribute of the new data which
enables the resulting matching.
Algorithm 2: Metadata matching.
1: Data.Frame ← read −CSV(old. f ile)
transform the dataframe to a two column data frame of attributes and
values
2: Data.Frame ← Re − map(Data.Frame)
Delete NULL values
3: Data.Frame ← Drop.Null.Values
4: Dictionary ←[]
5: i ← 0
6: for attribute in data f rame.attributes do
7: add [attribute : i] to Dictionary
8: i ← i + 1
9: end for
We replace the attributes by numerical values
10: Data.Frame[attributes] ← Data.Frame[Dictionary]
11: Labels ← Data.Frame[at tributes]
12: Vocab ← words in Data.Frame[values]
13: Vocab.int ← to − int (Vocab)
We create a matrix of data in numerical values
14: Feature ← matr ix(Vocab.int(Data.Frame[values]))
We split the data to training set and test set
15: train
x
,test
x
,train
y
,test
y
← train
t
est
s
plit( f eatures, labels)
16: train
y
,test
y
← one.hot.encoding(train
y
,test
y
)
17: model ← model.train(train
x
,train
y
)
18: Precision ← model. precision
19: New.Data ← Data.change from List.changes
20: Prediction ← model. predict(New.Data)
return Prediction
In terms of data processing, we transform all data
to strings. In the neural network, we use an embed-
ding layer, which purpose is compressing the feature
space of the input into a smaller one.
We use afterwards layers of LSTM (long short-
term memory) to conceptualize semantics in data, and
CNN (Convolutional neural network) layers to well
extract features of data.
For optimisation purposes (time execution and
precision), we decided to train the model on the at-
tributes (classes) changed. The predictions made will
be therefore applied to update the mappings.
3.1.5 Mapping File Update
In the integration process, the generated output is a
file of mappings connecting the metadata of the data
source with the ontology concepts. Our aim is to
change this file according to the predictions made by
the previous algorithm.
We perform this by replacing the changed at-
tributes in the mapping file, with the new ones pre-
dicted by the metadata matching algorithm.
4 IMPLEMENTATION AND
RESULTS
We present in this section the execution of our solu-
tion, as well as it’s application on different use cases.
4.1 Tools of Development
To manage the implementation of our approach, we
used the following tools :
• Google Colab
2
: We used Google colab as a
python platform to develop the proposed algo-
rithms and to build the prediction models of the
system.
• Keras
3
: We used the keras library to create, train
and test the prediction models of our system.
• Prot
´
eg
´
e
4
: An open source ontology editor and
a knowledge management system. We used it to
explore the ontology of drugbank available online.
4.2 Evaluation Metrics
To evaluate the classification models, we used the
confusion matrix. This latter is a table used to de-
scribe the performances of a classification model on a
set of data. It differentiates between the true and false
positives and negatives. It allows us to compute the
metrics of Precision, Recall, F-mesure and Accuracy.
TP / TN: True Positive / True Negative.
FP / FN: False Positive / False Negative.
• Accuracy : It’s the ratio between the total correct
predictions and the total predictions. Accuracy =
T P+T N
T P+T N+FP+FN)
2
Google Colab, https://colab.research.google.com
3
Keras, https://keras.io
4
Prot
´
eg
´
e, https://protege.stanford.edu
Semantic-based Data Integration and Mapping Maintenance: Application to Drugs Domain
473

• Precision: Proportion of well classed elements in
a particular class.
Precision =
T P
T P+FP
• Recall: Proportion of well classed elements over
the number of elements of a given class. Recall =
T P
T P+FN
• F1-score: Mesures the compromise between the
precision and the recall.
F1 −score =
2∗(Recall∗Precision)
(Recall+Precision)
Regarding the model’s calculated loss, we use the
Mean Square Error, to determine how much is the dif-
ference between the actual values and the predicted
values: y (Actual values), y
p
(Predicted values).
Loss = MSE =
1
n
∗
∑
n
i=1
(y
i
+ y
p
i
)
2
4.3 Classification Model
As for the classification model, we use two architec-
tures and compare the results. Te first architecture
depends on LSTM layers and the second depends on
CNN layers. The figure 4 presents these architectures.
We explore 2 use cases: 4 classes (4 attribute
changes) and 6 classes (6 attribute changes). We run
our approach in these cases, and capture the results.
4.4 Study Cases
The approach, and specifically the processing compo-
nent starts by detecting changes in files metadata. It
works by comparing lists of strings corresponding to
the metadata. The result is the list of changes with
attributes of the old file.
For the drugbank data we’re working with, we
have 55 attributes in the metadata. To illustrate the
result of the algorithm, we display changes over 10
attributes shown in table 1.
Table 1: Metadata change.
Old metadata drugbank-id name description cas-number unii
state groups general-references synthesis-reference indication
New metadata drug name specification cas-number unii
state groups general-references source indication
As we can see, some attributes have been changed
(colored attributes), others did not. The algorithm
returns the list of attributes that have been changed
[drugbank-id, description, synthesis-reference]. We
train therefore the models, and we present the results
as study cases.
4.4.1 Case of 4 Classes
We plot the training-validation accuracy and loss
throughout the whole training of the model. These
(a) LSTM based model
(b) CNN based model
Figure 4: Classification models.
plots (figure 6 a/b) describe the training phase of both
models. This phase has a training sub-phase in blue,
and a validation sub-phase in orange. We can see that
the LSTM model has a steady training accuracy, but
in contrast, it has a growing loss regarding the valida-
tion.
As for the CNN model, even though it lacks con-
sistency, it shows better results over the loss and ac-
curacy in training.
But these results do not fully represent the models
capacity, that’s why we test (test phase) them on a test
dataset, to compute the actual accuracy and recall.
Using the corresponding confusion matrices, we
compute the precision, recall, f1-score and accuracy
in table 2.
The test results show that, although the fact that
the LSTM model struggled and had poor results in
training, it has performed well. This is annotated by
ICSOFT 2022 - 17th International Conference on Software Technologies
474
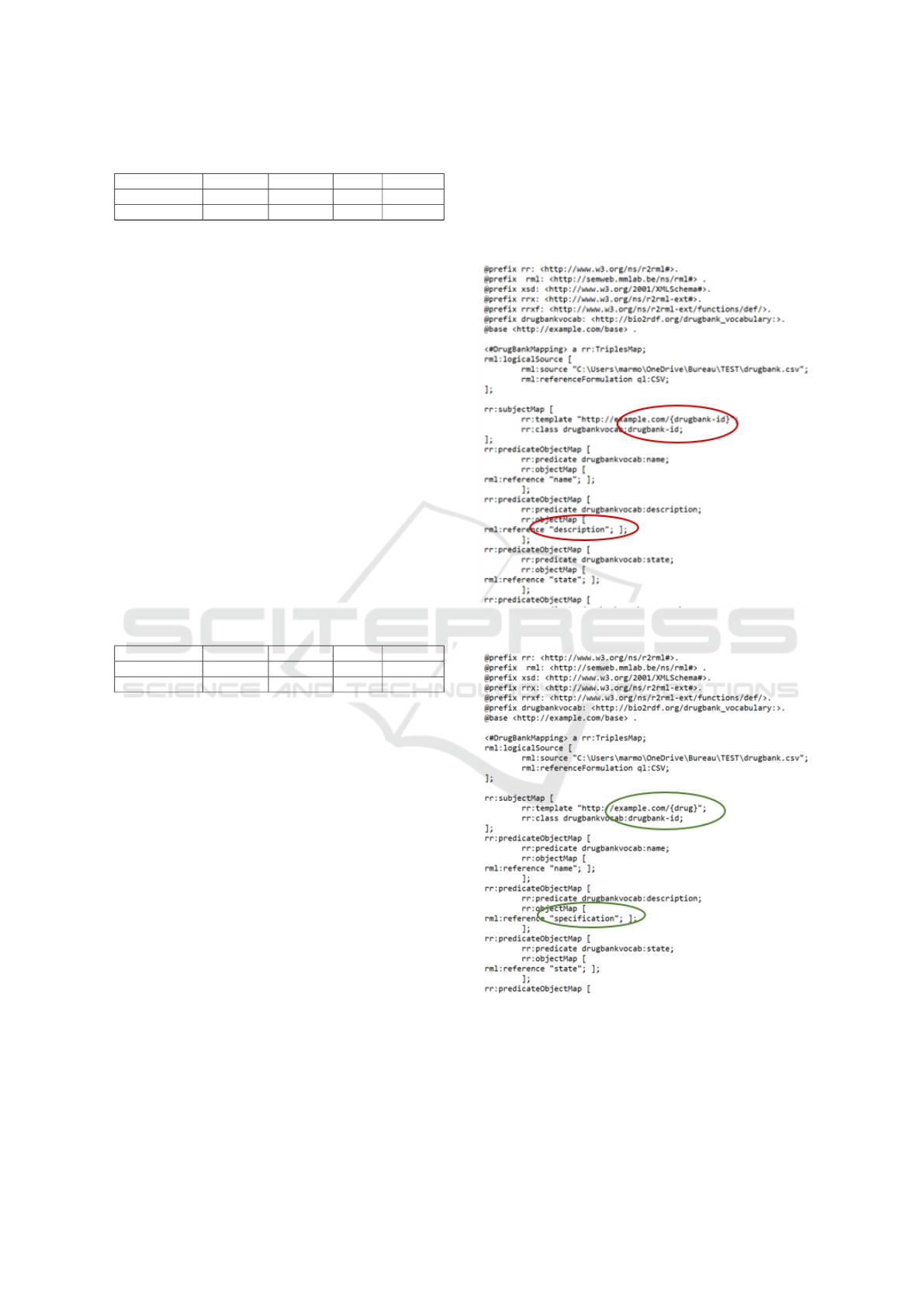
Table 2: Metrics of the models for 4 classes.
Model Accuracy Precision Recall F1-score
LSTM Model 0.93 0.859 0.855 0.86
CNN model 0.90 0.80 0.80 0.81
the accuracy reaching 93%.
But overall, both models got good results by
reaching an accuracy of 90%
4.4.2 Case of 6 Classes
We plot again the training-validation accuracy and
loss throughout the whole training of the model (fig-
ure 6 c/d).
The plots show that both models do train well, but
have trouble to well predict the classes in the valida-
tion. As validation loss, once again, the CNN model
show better results, but it lacks consistency.
We execute therefore the models on a test dataset
and compute evaluation metrics as shown in table 3.
As test results, LSTM model gives better perfor-
mance, even though it lacked a bit in the training
phase. These good results are due to it’s consistency
in the training, which is not the case for the CNN
model, who, the more it loses consistency, the more
poorly it performs.
Table 3: Metrics of the models for 6 classes.
Model Accuracy Precision Recall F1-score
LSTM Model 0.93 0.891 0.894 0.89
CNN model 0.85 0.74 0.72 0.70
But in general, the performance of both models is
good, by attending 85% for the CNN model and 93%
for the LSTM model.
4.4.3 Optimization
For both models, we have observed that they suffer
from an over-fitting problem. This problem means
that the model has a bad generalisation (Brownlee,
2018). It has learned too much that it performed well
on the training but poorly on a new data (test data).
So, to overcome this issue, we have applied some
regularisation techniques throughout our process to
get better results. Specifically, we used dropout and
early stopping.
By using both techniques, we managed to raise the
accuracy of both models to nearly 80%.
4.5 Mappings Update
As previously said, the classification algorithm takes
as input the data corresponding to the changes dis-
covered, and it outputs the attribute (class) of each set
of data. To update the mappings file, we take list of
changes and update the file correspondingly.
The final updated file (shown in figure 5) will en-
sure that the data integration approach works without
any issues, regardless of the metadata change of data
sources.
(a) Old mappings
(b) Updated mappings
Figure 5: Results of the mappings update.
Semantic-based Data Integration and Mapping Maintenance: Application to Drugs Domain
475
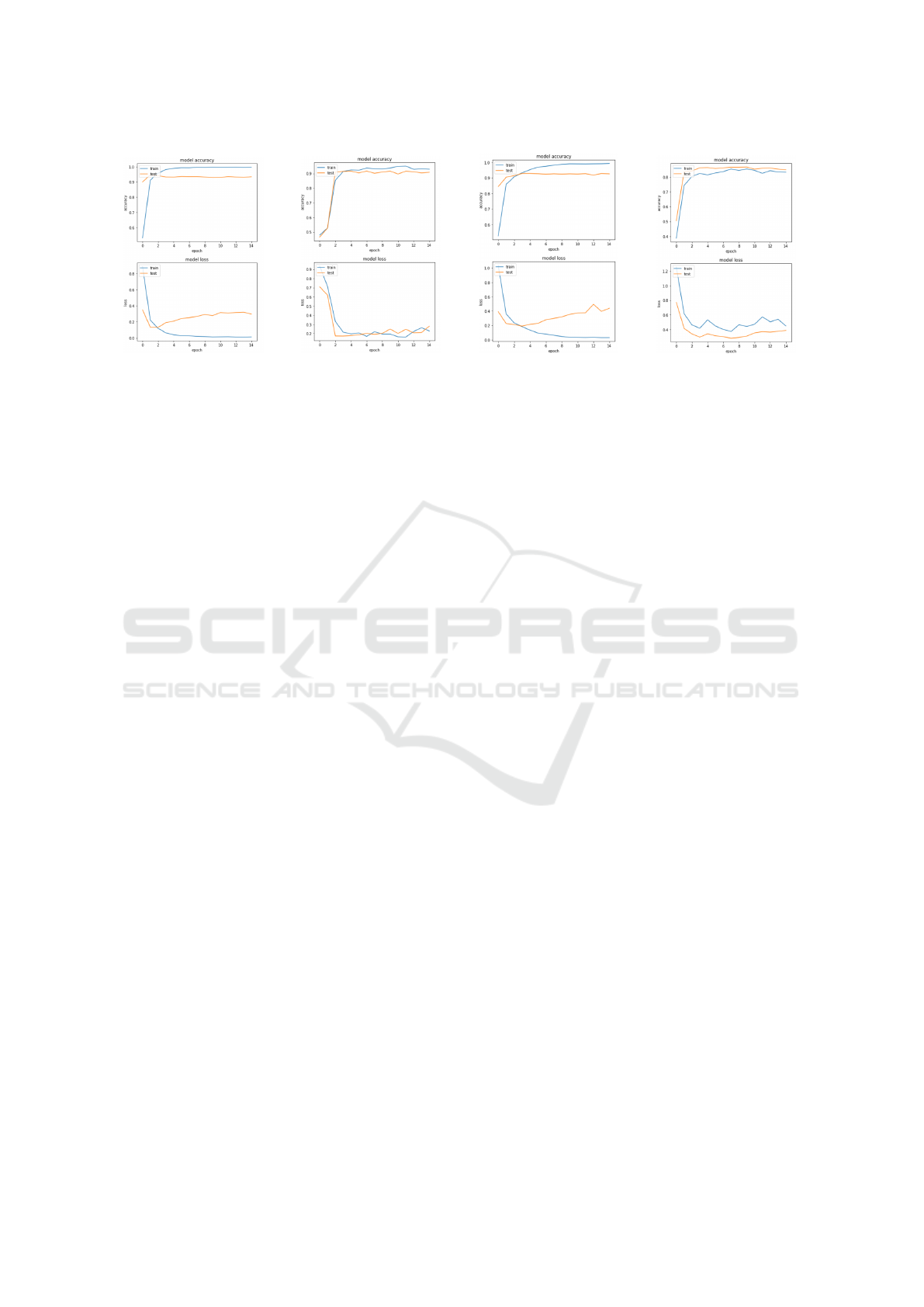
(a) LSTM based model
(4 classes)
(b) CNN based model (4
classes)
(c) LSTM based model
(6 classes)
(d) CNN based model (6
classes)
Figure 6: Test cases’ models training.
5 CONCLUSION AND
LIMITATIONS
In this paper, we give a response to the problem of
how to ensure the maintenance of the virtual data in-
tegration and metadata changes. Indeed, we present a
semantic-based data integration and mapping mainte-
nance approach with an application to drugs domain.
The proposed approach contains two main compo-
nents: a virtual semantic data integration component,
and a mapping update component. Both of them en-
sure the fully automated mapping maintenance of the
data integration. The proposal includes deep learning
techniques for the mapping maintenance. We have
performed our solution on two use cases, 4 and 6
changes in the data sources’ metadata. In each case,
we detect changes, and train a deep learning model to
classify the corresponding attribute based on its’ data.
We used also two types of models, an LSTM-based
model and a CNN-based model.
In terms of results, the models showed interest-
ing performances, as the accuracy reached 90%. Al-
though, the models did suffer from an over-fitting
problem, which is due to the similarity between the
different classes (attributes) in the dataset. Overall,
our solution is fully automated, which represents huge
benefits compared to the existing solutions that re-
quire human intervention. In matters of time execu-
tion, training models do not surpass the limit of 10
minutes for the worst-case scenario. This time exe-
cution is indeed long for an automated system, but
compared to the state of the art (human intervention),
it shows significant optimal result. We are currently
working on the number of classes that the model
needs to deal with. Indeed, as we handle data sources
changes, the data within these sources doesn’t contain
high rate of change. So, basically, the models work
on the same data that they were trained on, which will
enable high performance results. We intend also, to
manage not only one-to-one changes (one class corre-
sponding to only one class), but one to many changes,
which will improve the mapping maintenance. We
also plan, to improve our proposal to support differ-
ent multimodal data.
ACKNOWLEDGMENT
This work was funded by CY Initiative of Excellence
(grant ”Investissements d’Avenir” ANR- 16-IDEX-
0008), MuseMed Emergence project.
REFERENCES
Ahmad Abdullah Alqarni, E. P. (2017). Xml schema match-
ing using early dissimilarity detection approaches. In
La Trobe University Melbourne, Australia.
Brownlee, J. (2018). Better deep learning, train faster, re-
duce overfitting, and make better predictions.
Cesar, J. and al (2015). Dykosmap a framework for map-
ping adaptation between biomedical knowledge orga-
nization systems. In Journal of Biomedical Informat-
ics (EDBT).
Cesar, J. and al. (2015). State-of-the-art on mapping main-
tenance and challenges towards a fully automatic ap-
proach.
Do, H. (2007). Schema matching and mapping-based data
integration: architecture, approaches, and evaluation.
Khattaka, A. and al. (2015). Mapping evolution of dynamic
web ontologies. In Information Sciences.
Maria-Esther, V. and al. (2019). Semantic data integration
of big biomedical data for supporting personalised
medicine.
ICSOFT 2022 - 17th International Conference on Software Technologies
476

Masmoudi, M. and al (2021). Knowledge hypergraph-based
approach for data integration and querying: Applica-
tion to earth observation.
Mehdi, O. and al (2015). An approach for instance based
schema matching with google similarity and regular
expression. In University Putra Malaysia, Malaysia.
Mohamed-Amine, B. and al (2017). Schema inference for
massive json datasets. In The International Confer-
ence on Extending Database Technology (EDBT).
Munir, S. and al (2014). An instance-based schema match-
ing between opaque database schemas. In Proceed-
ings of the 4th International Conference on Engineer-
ing Technology and Technopreneuship (ICE2T).
N. Popitsch, B. H. (2018). Dsnotify – a solution for event
detection and link maintenance in dynamic datasets.
In Journal of web semantics.
Sahay, T. and al (2019). A schema matching using machine
learning. In 7th International Conference on Signal
Processing and Integrated Networks.
Semantic-based Data Integration and Mapping Maintenance: Application to Drugs Domain
477
