
CRISPR-Cas9: Components and Application
Jiawen Jiang
Department of Molecular and Cell Biology, University of Connecticut, 82 N. Eagleville, Mansfield, U.S.A.
Keywords: CRISPR-Cas9 System, sgRNA, crRNAs, Gene Editing, DNA Endonuclease, RuvC, HNH.
Abstract: The identification of components of the CRISPR system discovered in bacterial cells enables gene editing in
a more efficient way. The further upgrading allows researchers to easily edit the DNA sequence not only in
prokaryotic cells but also in mammalian cells. The novelization of the CRISPR-Cas9 system promotes broader
utilization of this technique, resulting in staggered cuts in dsDNA sequences. Even during the COVID-19
pandemic, the employment of CRISPR-Cas9 techniques accelerates the development of SARS-CoV-2 testing
kits, allowing fast and effective testing available to a massive population. In this paper, I describe the progress
of finding the components followed by the novelization as well as applications of the CRISPR-Cas9 system.
1 INTRODUCTION
1
An impassioned discussion about the CRISPR
(Clustered Regularly Interspaced Short Palindromic
Repeat) system has been initiated because of the
Nobel Prize winner Emmanuelle Charpentier and
Jennifer Doudna, the pioneers of CRISPR
technology, in 2020 (Savić, Schwank 2016). Early in
1987 when CRISPR has been discovered for the first
time, a sequence located in the bacterial genome that
disarms bacteriophages infection by cutting off the
phage’s DNA (Doudna, Charpentier 2014). This
bacterial adaptive immune system was first being
used for genome editing in 2012 when Charpentier
and Doudna thoroughly discover the feature of the
CRISPR-Cas9 system which can recognize a specific
sequence of DNA and cut off the target site (Savić,
Schwank 2016). The nuclease Cas 9 followed by two
guide RNA sequences together constitute the genetic
scissor (Savić, Schwank 2016). Since then, they were
also able to target different DNA sequences by
reprograming the sequence of the short RNA
(Doudna, Charpentier 2014). Scientists are now able
to genetically modify the sequence of short RNA and
insert that RNA inside Cas 9 protein to target and
disable the desired sequence of DNA in cells (Zhang
et al. 2014). Engineering not only the sequence of the
guide RNA but also the protein Cas 9 enable scientists
to add, delete, disable, enhance, or even replace the
sequence of interest much more easily than other
a
https://orcid.org/ 0000-0002-5449-2632
genome editing techniques such as Zinc Finger
Nuclease (ZFN), and Transcription-Activator Like
Effector Nucleases (TALEN) (Gupta et al. 2019).
This precious research achievement brings not
only people in the science field, but ordinary people’s
attention towards the application, advantages,
challenges, and potential shortcomings of this
technique. Thousands of questions have been
delivered to scientists to answer but some of them
have not yet been responded systematically.
In this review, with the aim of collectively and
comprehensively discussing why and how the
CRISPR-Cas 9 system deserves this massive
attention, the content of introduction of the CRISPR-
Cas 9 system, potential applications of CRISPR
technology, the progress of cutting-edge technology,
and the future opportunities of CRISPR-Cas9 will be
included. The significance of the objective focuses on
providing a thorough introduction of the CRISPR-
Cas9 system as well as its up-to-date application to
promote learning, thinking, and potential
breakthrough within this field.
744
Jiang, J.
CRISPR-Cas9: Components and Application.
DOI: 10.5220/0011294300003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 744-748
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
2 CRISPR-Cas9 SYSTEM
2.1 The Understanding of
CRISPR-Cas9
CRISPR-Cas9 was first understood by Jennifer
Doudna and Emmanuelle Charpentier in 2012 (Jinek
et al. 2012). Before 2012, Dr. Doudna worked on
understanding and analyzing the composition of the
CRISPR-Cas9 system which has been found out to be
involved in the bacterial innate immune system to
defend against viral infection (Wiedenheft et al.
2009). Working along with Dr. Charpentier, Dr.
Doudna identified the inner component of Cas9
protein consisted in the type II CRISPR-Cas9 system
(Hryhorowicz et al. 2017).
Cas9 is a DNA endonuclease guided by two
RNAs, one is called CRISPR RNAs (crRNAs) and
the other is trans-activating crRNAs (tracrRNA)
(Jinek et al. 2012, Cong et al. 2013). crRNA is
complementary to the sequence that Cas9 is targeting,
while tracrRNA is essential to crRNA maturation as
well as triggering plasmid cleavage (Jinek et al. 2012).
A specific motif appears in the foreign genome called
protospacer adjacent motif (PAM), 2-6 nucleotides
downstream of the cut site, also need to be recognized
by crRNA, and cut by Cas9 3-4 nucleotide upstream
of it (Jiang, Doudna 2017). Once part of the sequence
is fully recognized, Cas9 protein bind onto the strand,
making the cut using two domains, each domain cut
one DNA strand (Jinek et al. 2012) (Fig. 1a). By
utilizing radioactivating tags on one of the DNA
strands, Doudna et al. determined that the HNH
domain is responsible for complementary strand
cleavage, while the RuvC-like domain is for the
uncomplimentary strand cleavage (Jinek et al. 2012).
A linear dsDNA is obtained after the cleavage
showing a successful cut by Cas9 (Jinek et al. 2012).
Although this is a part of the innate immune
system of multiple bacteria such as Streptococcus
pyogenes, the research found out that the CRISPR-
Cas9 system is programmable and can be used in
other organisms by changing the guide RNA (Chen et
al. 2019). Moreover, crRNA and tracrRNA can be
linked together with a hairpin to become a chimeric
RNA that mimics crRNA: tracrRNA complex (Chen
et al. 2019, Ran et al. 2013). Studies show that by
editing the sequence of chimeric RNA, Cas9 protein
with a single chimeric guide RNA is potentially
capable of cutting any dsDNA in many organisms,
resulting in a new era of genomic regulation and
genomic editing (Chen et al. 2019, Ran et al. 2013).
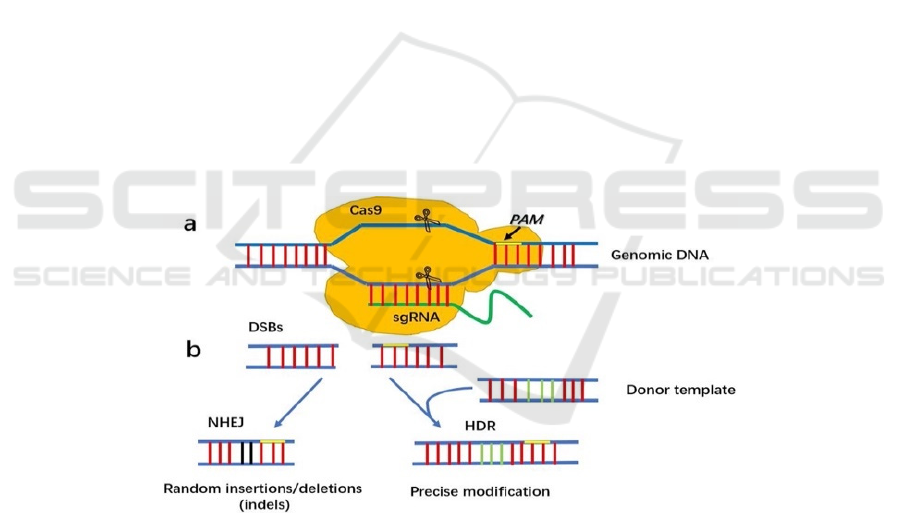
Figure 1. The CRISPR-Cas9 system. a. Double strand DNA breaks and binds to Cas9 and single guided RNA. Each domain
cut one strand of the DNA and resulting in double-strand break of genetic material (Chen et al. 2019). The original circular
DNA will become linear DNA due to the cut (Jinek et al. 2012). b. The double-strand lesions will be repaired through either
a non-homologous end joining or a homologous repair pathway. Both NHEJ and HDR allow the desired sequence to be
eliminated from the DNA achieving the goal of attacking foreign DNA (Chen et al. 2019, Ran et al. 2013, Cong et al. 2013).
2.2 Repurposing CRISPR-Cas9 System
The CRISPR-Cas9 system is programmed not only to
cut dsDNA but also to manipulate transcription in
eukaryotes (Qi et al. 2013). Inactive Cas9 or
denatured Cas9 (dCas9) protein is a great model in
the experiment (Cong et al. 2013) (Fig. 2). In this case,
the Cas9 lacking the endonucleolytic activity can
normally bind to complementary strand DNA but
cannot cut desired sequence [Qi et al. 2013]. Research
has shown that while Cas9 binds to the dsDNA, the
transcription is blocked with low off-target effects (Qi
et
al. 2013, Hsu et al. 2014). Furthermore, Cas9 has
CRISPR-Cas9: Components and Application
745
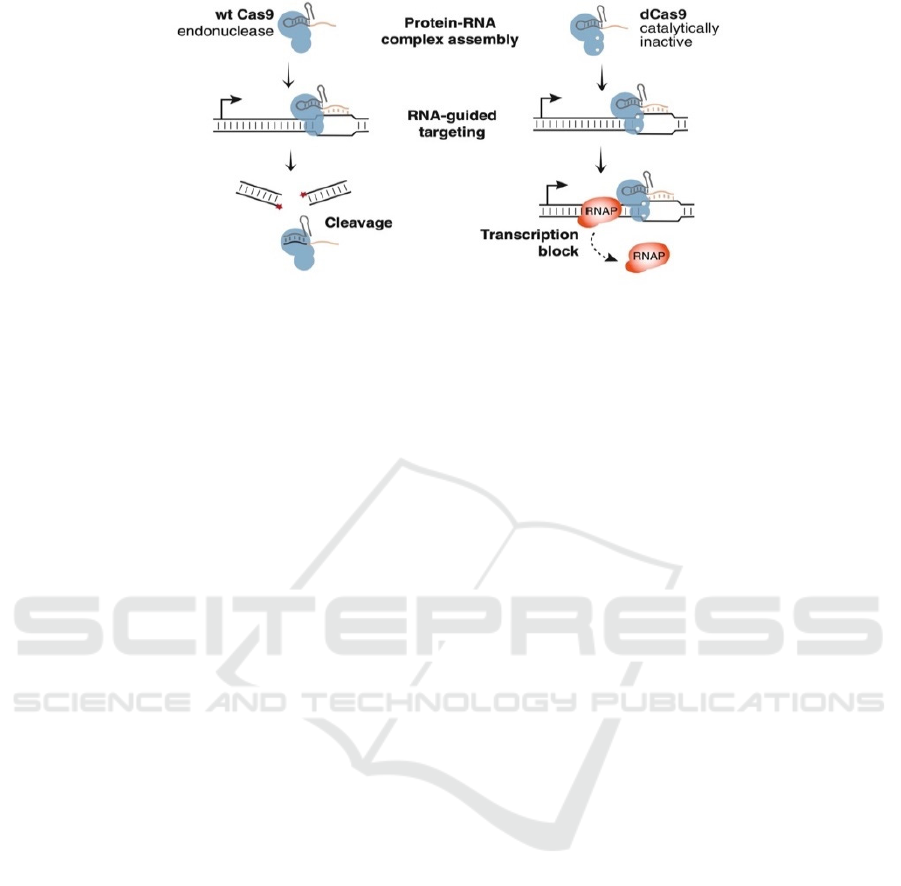
Figure 2. The programmable Cas9 protein and single-guide RNA (sgRNA) enable transcription regulation (Qi et al. 2013).
The wild type Cas9 protein cut results in the forming of linear DNA (left), while a dead Cas9 protein with a modified sgRNA
can bind onto the DNA sequence and shut down the original transcription by occupying the transcriptional site where the
RNA polymerase (RNAP) needs to bind (right) (Qi et al. 2013).
been used in multiple eukaryotic organisms to test the
efficiency and accuracy of genetic regulation and as a
result, the outcome is positive in most of the
organisms (Hsu et al. 2014).
The system originated from bacteria is used to cut
off and silence viral DNA (Jinek et al. 2012, Ran et
al. 2013). Silencing of the desired segment of dsDNA
in eukaryotic cells is achieved by cutting off the target
sequence followed by adding more nucleotide base
pairs to close the gap (Ran et al. 2013). Cas9 cutting
resulting in target double-strand DNA breaks induces
non-homologous end-joining or homologous repair
pathway (Chen et al. 2019, Cong et al. 2013, Hsu et
al. 2014) (Fig. 1b). Both pathways contribute to
precise gene editing by silencing a specific gene
transcription with very low off-target effects.
Furthermore, research carried out in mammalian cells
indicated that the CRISPR-Cas9 system could also
show high efficiency of creating target DNA lesions
with the desired sgRNA (Hsu et al. 2014).
Cong et al. contributed significantly to the
understanding and further investigation of the type II
CRISPR-Cas9 system in human cells (Cong et al.
2013). Human codon-optimized Streptococcus
pyogenes Cas9 (SpCas9) and RNase III (hSpRNase
III) are modified versions of Cas9 and RNase that can
be better used in human cells and at the same time
have less error with higher efficiency (Cong et al.
2013).
According to Cong et al., the type II CRISPR-
Cas9 system can be as efficient and accurate as of the
TALENs which has been used in genetic editing prior
to the discovery of CRISPR technology (Cong et al.
2013). Furthermore, they claimed that CRISPR-Cas9
system can do multiplexed genome engineering
(Cong et al. 2013). Cas9 is engineered to target both
EMX1- and PVALB loci. The efficiency is 1.6%,
indicating the ability to do multiplexed editing in a
single genome (Cong et al. 2013). The efficiency of
cutting is diminished by mismatches between the
sgRNA and target DNA (Slaymaker et al. 2015).
Neutralization of positively charged groove between
HNH, RuvC, and PAM interacting domains on the
nontargeting DNA strand promotes rehybridization
between dsDNA strands and hence encourages tight
binding between sgRNA and target DNA strand
(Slaymaker et al. 2015). This upgrade by Skaymaker
et al. allows CRISPR-Cas9 system to reach a newer
level with a very low off-target effect in terms of
target cutting (Cong et al. 2013, Slaymaker et al.
2015).
2.3 Novelizations and Progression of
the Application of CRISPR-Cas9
System
More modifications on the Cas9 system by Zetsche et
al. enabled more various binding and cutting. For
example, a novel version of Cas9 called Cpf1 and
C2c2 endonuclease achieved a staggered cut on
dsDNA (Zetsche et al. 2015).
Another significant update on CRISPR-Cas
system by Gootenberg et al. was using the method of
specific high-sensitivity enzymatic reporter
unlocking (SHERLOCK) to detect foreign strains in
small amounts such as the infection by Ziska and
Dengue virus (Gootenberg et al. 2017). The
advantage of this technique is that it can sense viral
and bacterial pathogens in samples that are relatively
advisable such as human serum or urine samples
(Gootenberg et al. 2017). Due to the high accuracy of
the technique, it can distinguish between similar viral
or bacterial strains successfully (Gootenberg et al.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
746

2017). It can also distinguish SNP in human
genotyping (Gootenberg et al. 2017).
During the COVID-19 pandemic, Joung et al.
discovered STOPCovid, a technique used to rapidly
detect the genetic material of SARS-CoV-2 from oral
or nasal swab samples giving either positive or
negative outcomes using detection strips or specific
values of viral RNA counts under fluorescence
readers (Joung et al. 2020). The overall sensitivity is
93.1% and specificity is 98.3% among 402 candidates
(Joung et al. 2020).
The rapid, inexpensive, and sensitive detection
enables CRISPR-Cas system to be highly used since
2015 in genetic regulation and genomic editing
(Gootenberg et al. 2017, Joung et al. 2020).
Recently, studies have shown that the success of
CRISPR-Cas9 technology inspired therapy in
Huntington’s disease, sickle cell diseases, cancer, and
B-thalassemia, indicating more potential treatments
available in the future targeting different diseases
(György 2021).
3 DISCUSSION
Owing to the concern of creating genetic mutation
due to permanent change in genes in human cells by
the CRISPR-Cas9 technology, it is better having
safely regulate the on-and off-switches of the
CRISPR-Cas9 system (Shivram et al. 2021). In case
of observing relatively high off-target effect that may
lead to serious genetic defects as well as
unpredictable diseases of an organism, having types
of “emergency shut down system” is necessary for
more complicated gene editing events, especially
when introducing DNA cut and gene segment
replacement in mammalian cells or even in human
cells. Regulators from Escherichia coli and
Salmonella typhi are identified as having H-NS and
LRP which are responsible for negative regulation of
Cas expression, while LeuO can positively regulate
Cas promoter to further simulate CRISPR-Cas cutting
(Shivram et al. 2021). Those genes, in theory, are
responsible for regulating Cas system, as well as
being potential switches for CRISPR-Cas9 system,
but the future investigation is needed to make sure
those factors found in bacteria can be equally
beneficial with high efficiency in mammalian cells.
Moreover, even though the off-target effect is
significantly lowered by Zhang et al. after several
steps of upgrading, it is not possible for now to achive
0 off-target effects and as a result, it is still unknown
if the efficiency can promote a positive outcome in
clinical trials (Cong et al. 2013, Zetsche et al. 2015).
4 CONCLUSION
The existence of components located in CRISPR-
Cas9 system allow further investigation and
advancement in other fields. The revelation of the
ability to practice precise cut by RuvC-like and HNH
domain, engineer sgRNA, and replicate the
mechanism in eucaryotic cells results in a higher
frequency of the usage of the CRISPR-Cas9
technique in labs regarding the aim of doing gene
editing. This technique is known as the most rapid
and the easiest method in genetic regulation and
genomic engineering. Not only the Noble Prize
winner Emmanuelle Charpentier and Jennifer
Doudna but also other scientists progressively and
largely contribute to the refinement of the CRISPR-
Cas9 system in their research fields. Based on all of
the existing gene editing products by the CRISPR-
Cas9 system, investigating an efficient way of fully
controlling the on and off of CRISPR-Cas9 is critial
as well as worth studying in order to make CRISPR-
Cas9 a precise gene editing machinery. This paper
about CRISPR-Cas9, an influential topic in the
science field, hopefully, will promote thinking as well
as more discoveries that may lead to better and novel
disease treatments in the future.
ACKNOWLEDGEMENT
I would like to extend my sincere thanks to Dr.
Gerwald Jogl at Brown University for providing
advice on how to improve this paper.
REFERENCES
Chen, M., Mao, A., Xu, M., Weng, Q., Mao, J., and Ji, J.
(2019) CRISPR-Cas9 for Cancer Therapy:
Opportunities and Challenges. Cancer Letters, 447: 48–
55.
Cong, L., Ran, F., Cox, F., Lin, S., Barretto, R., Habib, N.,
Hsu, P., Wu, X., Jiang, W., Marraffini, L., Zhang, F.
(2013) Multiplex Genome Engineering Using
CRISPR/Cas Systems. Science, 339(6121): 819–823.
Doudna, J. A., Charpentier, E. (2014) The new frontier of
genome engineering with CRISPR-Cas9. Science,
346(6213). https://doi.org/10.1126/science.1258096
Gootenberg, J., Abudayyeh, O., Lee, J., Essletzbichler, P.,
Dy, A., Joung, J., Verdine, V., Donghia, N., Daringer,
N., Freije, C., Myhrvold, C., Bhattacharyya, R., Livny,
J., Regev, A., Koonin, E., Hung, D., Sabeti, P., Collins,
J., Zhang, F. (2017) Nucleic Acid Detection with
CRISPR-Cas13a/C2c2. Science, 356(6336): 438–442.
CRISPR-Cas9: Components and Application
747

Gupta, D., Bhattacharjee, O., Mandal, D., Sen, M. K., Dey,
D., Dasgupta, A., Kazi, T. A., Gupta, R., Sinharoy, S.,
Acharya, K., Chattopadhyay, D., Ravichandiran, V.,
Roy, S., Ghosh, D. (2019). CRISPR-Cas9 system: A
new-fangled dawn in gene editing. Life Sciences, 232.
https://doi.org/10.1016/j.lfs.2019.116636
György, B. (2021) CRISPR Cuts Disease Course Short in
Blood Disorders. Science Translational Medicine,
13(575): eabg1756.
Hryhorowicz, M., Lipiński, D., Zeyland, J., Słomski, R.
(2017) CRISPR/Cas9 Immune System as a Tool for
Genome Engineering. Archivum Immunologiae et
Therapiae Experimentalis, 65(3): 233–240.
Hsu, P., Lander, E., Zhang, F. (2014) Development and
Applications of CRISPR-Cas9 for Genome
Engineering. Cell, 157(6): 1262–1278.
Jiang, F., Doudna, J.A. (2017) CRISPR–Cas9 Structures
and Mechanisms. Annual Review of Biophysics, 46(1):
pp. 505–529.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.,
Charpentier, E. (2012) A Programmable Dual-RNA-
Guided DNA Endonuclease in Adaptive Bacterial
Immunity. Science, 337(6096): 816-821.
Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel
M, Barretto RPJ, Ranu A, Macrae RK, Faure G,
Ioannidi EI, Krajeski RN, Bruneau R, Huang MW, Yu
XG, Li JZ, Walker BD, Hung DT, Greninger AL,
Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang
F.(2020) Detection of SARS-CoV-2 with SHERLOCK
One-Pot Testing. New England Journal of Medicine,
383(15): 1492–1494.
Qi, L., Larson, M., Gilbert, L., Doudna, J., Weissman, J.,
Arkin, A., Lim, W. (2013) Repurposing CRISPR as an
RNA-Guided Platform for Sequence-Specific Control
of Gene Expression. Cell, 152(5): 1173–1183.
Ran, F., Hsu, P., Weight, J., Aggrwala, V., Scott, D., Zhang,
F. (2013) Genome Engineering Using the CRISPR-
Cas9 System. Nature Protocols, 8(11): 2281–2308.
Savić, N., Schwank, G. (2016) Advances in therapeutic
CRISPR/Cas9 genome editing. Translational Research,
168. https://doi.org/10.1016/j.trsl.2015.09.008
Shivram, H., Cress, B., Knott, G., Doudna, J. (2021)
Controlling and Enhancing CRISPR Systems. Nature
Chemical Biology, 17(1): 10–19.
Slaymaker, I., Gao, L., Zetsche, B., Scott, D., Yan, W.,
Zhang, F. (2015) Rationally Engineered Cas9
Nucleases with Improved Specificity. Science,
351(6268): 84–88.
Wiedenheft, B., Zhou, K., Jinek, M., Coyle, S., Ma, W.,
Doudna, J. (2009) Structural Basis for DNase Activity
of a Conserved Protein Implicated in CRISPR-
Mediated Genome Defense. Structure, 17(6): 904–912.
Zetsche, B., Gootenberg, J., Abudayyeh, O., Regev, A.,
Kolonin, E., Zhang, F. (2015) Cpf1 Is a Single RNA-
Guided Endonuclease of a Class 2 CRISPR-Cas System.
Cell, 163(3): 759–771.
Zhang, F., Wen, Y., Guo, X. (2014) CRISPR/Cas9 for
genome editing: progress, implications and challenges.
Human Molecular Genetics, 23(R1).
https://doi.org/10.1093d/hmg/ddu125
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
748
