
Analyzing Zelda and Other Transcription Factors That Regulating
Ubiquitous and Patterning Genes of Drosophila Melanogaster
Shuyang Wang
School of Chemistry and Molecular Bioscience, Faculty of Science, University of Queensland,
Brisbane, St Lucia 4072, Australia
Keywords: Transcription Factor, Drosophila Melanogaster, Zelda, Ubiquitous (Class 1) Gene, Patterning (Class 2) Gene.
Abstract: The embryonic developments of many animals are controlled by a series of genes whose counterparts can be
found in human. Therefore, analyzing the molecular nature of these genes is important for extending
understanding of genetic diseases in human. In this research, Drosophila melanogaster was used as the model
organism to analyze how their genes are regulated by different transcription factors during early
developments. Each student was assigned 8 unique genes, their transcription factor binding was subsequently
analyzed with software and online platforms. A gene’s Zelda binding level can be monitored by IGB to
determine whether it’s expressed ubiquitously or regionally. The results also proved that patterning genes and
ubiquitous genes can regulate the transcription levels of each other.
1 INTRODUCTION
Transcription factors are proteins that bind to DNA
strands to control the rate of transcription, the process
in which DNA is used as template to produce RNA
that is required for subsequent production of proteins
(Lambert 2018). The DNA sequences are identical in
all cells of an individual organism. However, DNA
can only influence the organism when it is translated
into particular gene products, proteins. Therefore, the
DNA expression underpinned by sophisticated
transcription factor network determines everything
related to the phenotype and physiological functions
of the organism (Signor, Nuzhdin 2018).
In this project conducted by Professor Christine
Rushlow in New York University, students analyzed
the binding level of different transcription factors for
ubiquitous (Class I) and patterning (Class II) genes of
Drosophila Melanogaster embryos (Alberts 2002).
Drosophila is a very commonly used model organism
in biological science. It’s famous for the low cost and
efficient reproduction (Tolwinski 2017). Drosophila
genes can be assigned into 2 classes. Class 1 genes
have ubiquitous expression over the whole embryo.
Therefore, they are also known as ubiquitously
expressed genes. In comparison, Class 2 genes have
specific expression patterns, they are only expressed
at particular position of the drosophila embryo.
That’s why they are also known as patterning genes.
Zelda is an important transcription factor that
activates zygotic genes by binding to sequences with
TAG base pairs and making the chromatin accessible
for other transcription factors (Ventos-Alfonso, Ylla,
Belles 2019). Zelda is expressed ubiquitously in
embryo due to its overarching function for
transcription. However, its effects to Class 1 and
Class 2 genes are different. For Class 1 genes, it
directly binds to the promoter. Promoter is a DNA
sequence segment that works as the ‘switch’ that
initiating the transcription of the gene
(Mikhaylichenko et al 2018). If the Zelda binds and
turns it on, the gene will be transcribed into RNA
molecules that will further be translated into the gene
product, protein. Otherwise, if Zelda is knocked out
in experiments, no Class 1 genes can be transcribed
as the ‘switches’ are totally turned off. This is
different in Class 2 genes. Zelda will only binds to the
enhancer of the patterning genes to regulate their
transcription levels. Enhancer is a DNA sequence that
recruits different transcription factors to control the
rate of transcription (Zabidi, Stark 2016). Zelda
works as an activator and binds to the enhancer
regions of Class 2 genes to increase the transcription
levels.
Zelda doesn’t control all the transcription
processes by itself. In fact, expression of all genes are
Wang, S.
Analyzing Zelda and Other Transcription Factors That Regulating Ubiquitous and Patterning Genes of Drosophila Melanogaster.
DOI: 10.5220/0011297000003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 817-826
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
817

regulated by sophisticated transcription factor
networks. Different transcription factors are recruited
to the promoter and enhancer of a gene and act
synergistically to provide the optimal transcription
level of this gene for embryonic development. Zelda
as well as other important transcription factors are
what we are interested in. In this project, I’ve used
software and data from Professor Christine’s lab to
investigate the transcription regulation of some
typical Drosophila embryonic Class 1 and 2 genes.
Understanding the transcriptional regulation in
Drosophila will enable deeper understanding of
analogous processes in human, which may provide
some key ideas to develop therapies of hereditary
diseases.
2 METHODS
2.1 Integrated Genome Browser (IGB)
Integrated Genome Browser (IGB) is a software that
can use graphs to depict the extents of RNA
transcription and transcription factor binding for all
genes in Drosophila Melanogaster. It also shows us
the position of the genes by indicating the base pair
numbers in each chromosome.
The IGB software was opened. Next, the Species
Drosophila Melanogaster and the 2014 version of
genome was selected. Subsequently, Drosophila data
was then uploaded into the IGB. The data includes:
RNA transcription during 12 and 13 cycle of the
wildtype Drosophila embryonic development into
IGB. The 13 cycle RNA transcription of embryos
with Zelda knocked out. The Zelda binding level data
during the 8 embryonic cycle of the wildtype embryo.
The DNA sequence tag with high affinity to Zelda.
After typing the name or code of the specific gene
(e.g. CG1641/sisA), IGB software depicted the data
as graphs Figure 1, 2. The IGB graphs are used to
distinguish whether the gene belongs to Class 1 or
Class 2. They are also used for analysis in JASPAR
and NCBI websites later.
2.2 National Center for Biotechnology
(NCBI)
NCBI website (www.ncbi.nlm.nih.gov) (NCBI
website https://www.ncbi.nlm.nih.gov) is used to
find the DNA sequence at the promoter or enhancer
of specific gene. Choose ‘Gene’ at the search box,
then type the name or code of the gene and search.
Then click ‘FASTA’ to see the whole gene sequence
represented by nitrogen bases (A, T, C, G). The
website also tells us the region of the gene and in
which chromosome the gene is. Finding the nitrogen
base number of the Zelda binding peak shown in IGB
graphs, minus and plus 200 base pairs and click
‘update view’. Then NCBI will show us the 400bp
sequence with Zelda bound. Finally, the 400bp
sequence was copied and pasted into JASPAR
website to identify all transcription factors binding to
this sequence.
2.3 JASPAR Database
JASPAR (JASPAR website 2020), a website used to
find all transcription factors binding to specific
sequence in the genome. At first, Drosophila
Melanogaster was chosen as the organism model in
JASPAR. The 400bp DNA sequence generated in
NCBI was then pasted into JASPAR and scanned.
JASPAR would show all transcription factors bound
to that 400bp region as well as the affinity of binding
indicated by scores. Finally, the JASPAR data was
downloaded as CSV Transcription factors with
binding scores higher than 10 were chosen for
subsequent analysis. These extracted transcription
factors were shown in Table 1,2,3,4.
2.4 Berkeley Drosophila Genome
Project (BDGP)
BDGP website (https://insitu.fruitfly.org/cgi-
bin/ex/insitu.pl)(BDGP website) is used to find
expression patterns of Class 1 and Class 2 genes to
monitor where a particular gene is expressed in the
embryo Figure 5.
3 RESULTS
3.1 Integrated Genome Browser (IGB)
Results
According to the IGB results of Class 1 genes shown
in Figure 1 and patterning genes shown in Figure 2,
the transcription of Class 1 genes except
CG4261/term changed to 0 if Zelda is knocked out.
However, the transcription of patterning genes only
reduced instead of disappearing when Zelda was
knocked out.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
818
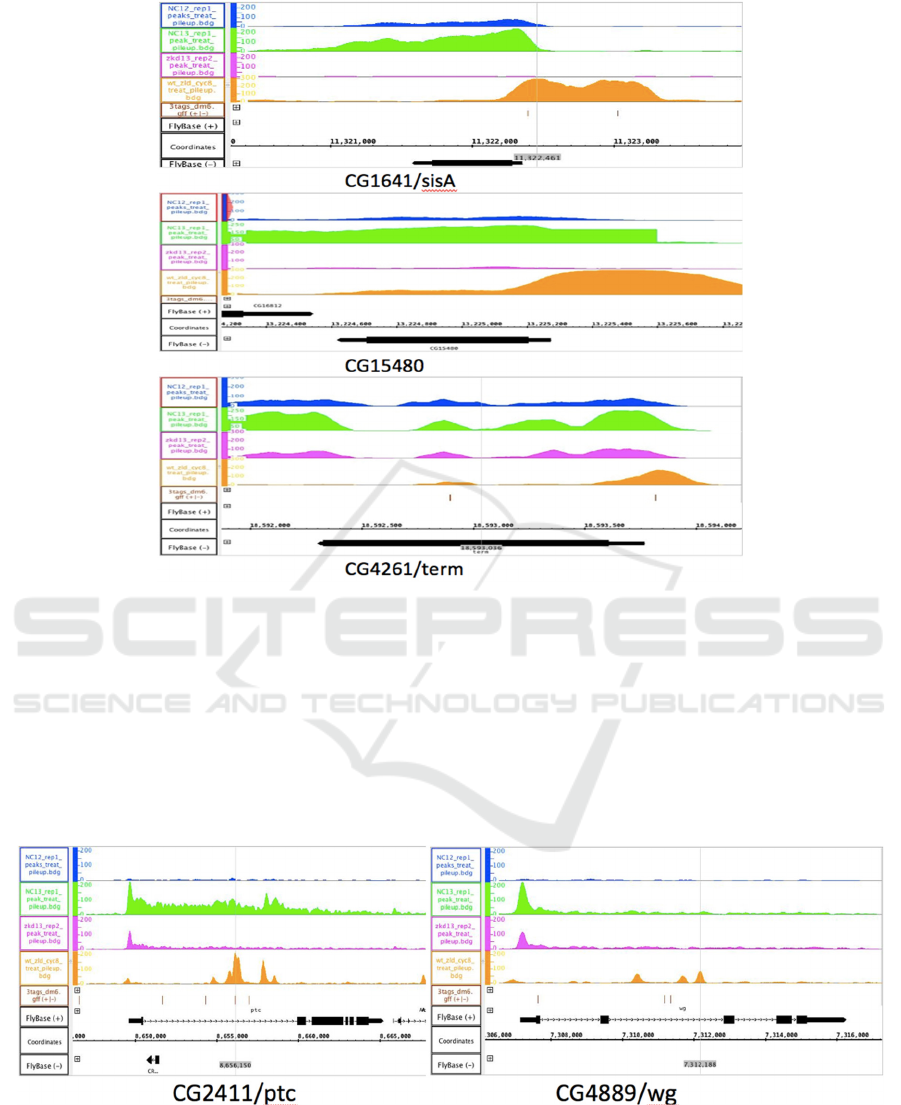
Figure 1: IGB results of Class 1 (ubiquitous) genes.
CG1641/sisA at Chromosome X, CG15480 at
Chromosome 2L and CG4261/term at Chromosome
3L. The X-axis shows the genomic position of the
chromosome in base pair. The black bar with an
arrow at one end represents the sisA gene and the
arrow shows the orientation of the gene. The blue and
green graph shows the RNA transcript levels in cycle
12 and cycle 13 respectively in wild type embryos of
Drosophila Melanogaster. The pink graph shows the
cycle 13 RNA level in fruit fly embryos whose Zelda
genes are knocked out. In contrast to these 3 graphs,
the orange graph reveals the level of Zelda binding,
instead of RNA. The Zelda binding tags,
CAGGTAG, are represented by the brown dashes
below the Zelda binding level graph. At the promoter
of gene sisA, 2 Zelda binding peaks were identified.
Figure 2: IGB results of patterning (Class2) genes.
CG2411/ptc at Chromosome 2R and CG4889/wg
at Chromosome 2L. All graphs represent the same
variables as mentioned in Figure 1. The dash line in
the gene black bar indicates the introns of the gene.
Analyzing Zelda and Other Transcription Factors That Regulating Ubiquitous and Patterning Genes of Drosophila Melanogaster
819
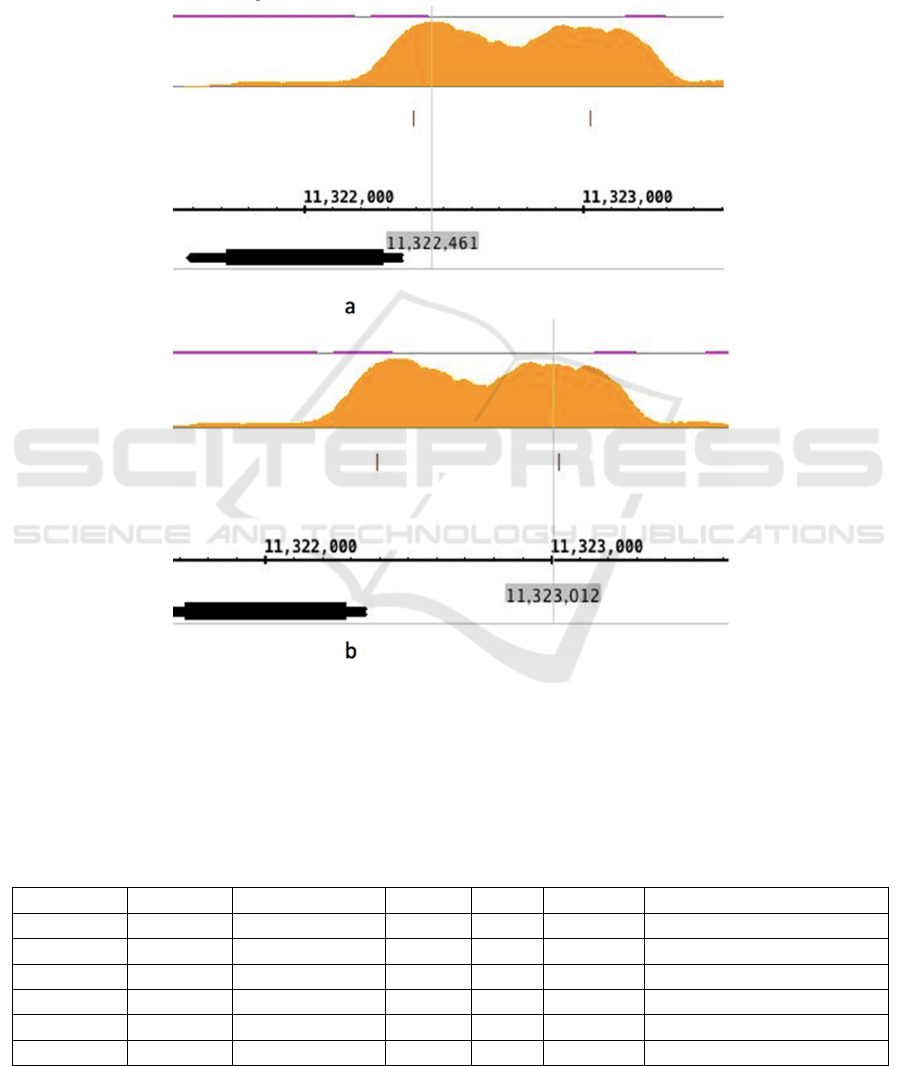
3.2 Transcription Factors Bound
around the Zelda Binding Peaks of
Class 1 Gene CG1641/Sisa
Figure 3. shows the Zelda binding levels at the
promoter region of CG1641/sisA in Chromosome X.
The transcription factor binding levels around the 2
Zelda binding peaks are shown by Table 1. and
Table 2. According to these two tables, Zelda (vfl)
and Kr bound at the highest level. 5 binding
sequences for Zelda and 4 binding sequences for Kr.
Kr binding regions are highly overlapped.
Transcription factors zen2, bcd, cad dl were also
found at high level around Zelda binding peak.
Figure 3: Zelda binding peaks at the promoter region of Class 1 gene CG1641/sisA.
This figure incorporates 2 screenshots of the IGB
graph result of the Class 1 ubiquitous gene
CG1641/sisA. 2 Zelda binding peaks are shown in a.
and b. respectively. The vertical grey lines show the
approximate Zelda binding peaks, together with grey
highlighted numbers representing the exact locations
of peaks in Chromosome X of Drosophila
melanogaster.
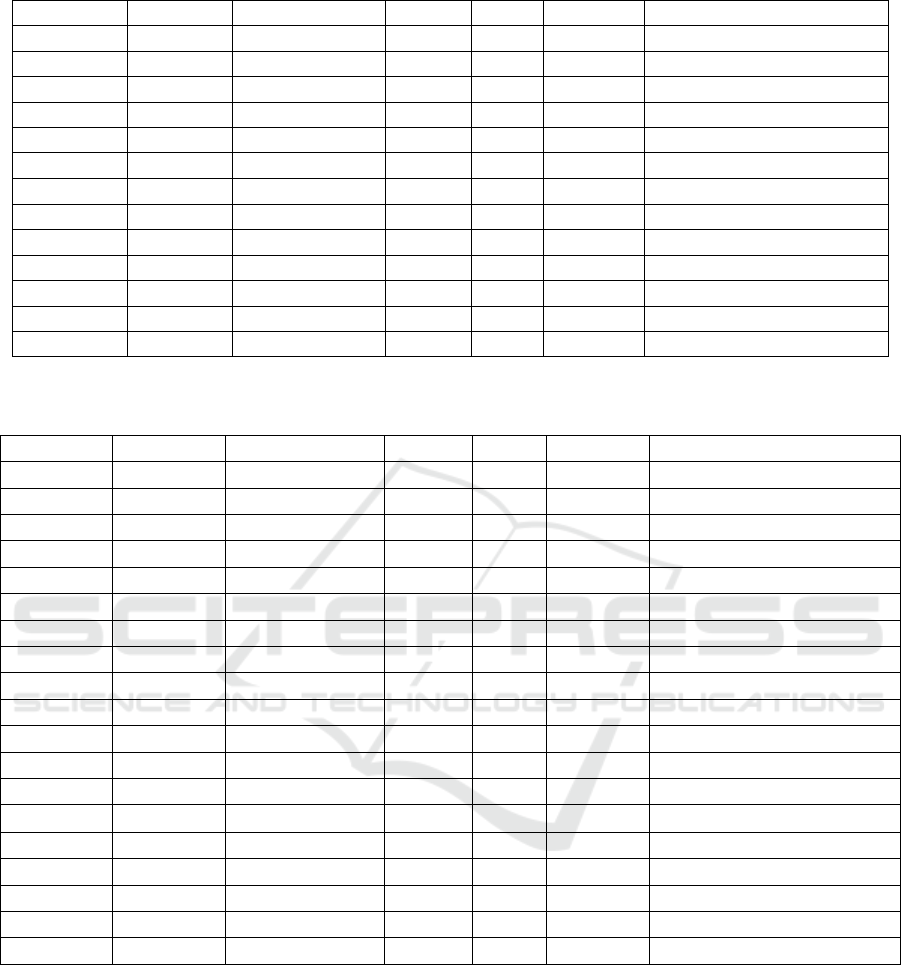
Table 1: Binding levels of different transcription factors around the Zelda binding peak 11322461 (Figure 3a.) in promoter
region of CG1641/sisA.
Name Score Relative score Start End Strand Predicted sequence
vfl 14.1057 0.99878905 139 150 + ATGCAGGTAGGC
slp1 13.9241 0.992220694 104 114 + TTGTTTACATA
vfl 13.6419 0.989526395 257 268 - CCGCAGGTAGCT
D 12.7559 0.943674382 41 51 + CCTTTTGTTTT
vfl 12.7267 0.971251085 66 77 + TATCAGGTAGAC
cad 12.3672 0.939626548 186 196 + GATCATAAATC
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
820
dve 12.1441 0.985360611 95 102 + CTAATCCC
fkh 12.0097 0.935520664 105 115 + TGTTTACATAT
D 11.61 0.915055081 100 110 + CCCTTTGTTTA
D
r
11.5272 0.999999983 61 67 + CCAATTA
K
r
10.8198 0.880272744 94 107 + CCTAATCCCTTTGT
br
10.7731 0.875449066 42 55 - TTTAAAAACAAAAG
sna 10.7449 0.886467281 30 42 - GGATCAGGTGCGA
sna 10.7444 1.000000017 33 38 - CAGGTG
K
r
10.703 0.889807779 96 106 - CAAAGGGATTA
nub 10.699 0.848601417 103 114 - TATGTAAACAAA
Ptx1 10.654 0.955186943 95 101 + CTAATCC
b
r(var.4) 10.651 0.925211296 103 113 - ATGTAAACAAA
b
cd 10.5191 0.999999998 96 101 + TAATCC
CG11617 10.4776 0.959704496 107 113 + TTTACAT
Table 2: Binding levels of different transcription factors around the Zelda binding peak 11323012 (Figure 3b.) in promoter
region of CG1641/sisA.
Name Score Relative score Start End Strand Predicted sequence
K
r
16.3492 0.96373581 16 29 + TTTAACCCTTTGAG
pn
r
14.8762 0.96410016 253 263 - TATCGATTGCC
BEAF-32 14.0397 0.94023386 256 270 - ACACCAATATCGATT
vfl 13.698 0.99064777 239 250 + TAGCAGGTAGCA
Dref 13.3379 0.95960864 256 265 - AATATCGATT
BEAF-32 13.0371 0.93733098 254 265 + GCAATCGATATT
BEAF-32 13.0371 0.93733098 254 265 - AATATCGATTGC
EcR::usp 12.9829 0.91055415 40 54 + CAGGTCGCTGAACCC
vfl 12.6211 0.96914093 176 187 - CATCAGGTAGCC
K
r
12.5593 0.93659564 18 28 - TCAAAGGGTTA
cnc::maf-S 12.2768 0.91497239 355 369 - AATGAGTCAACAAAT
Lim1 12.2032 1.00000002 79 85 + TTAATTA
Lim1 12.2032 1.00000002 80 86 - TTAATTA
al 12.1468 1 79 85 - TAATTAA
al 12.1468 1 80 86 + TAATTAA
zen2 10.5275 1 79 85 + TTAATTA
zen2 10.5275 1 80 86 - TTAATTA
b
cd 10.5191 1 115 120 + TAATCC
dl 10.1277 0.86361398 188 199 - GTGGGTTTCCCG
3.3 Transcription Factors Bound
around the Zelda Binding Peaks of
Class 2 Gene CG2411/Ptc
Distinguishable with Zelda binding in Class 1 gene
sisA, Zelda bound to the intron region of the
patterning gene ptc as shown in Figure 4. The Zelda
binding peaks in ptc are separate instead of merged
together like that in sisA Figure 3. Shown by Table
3. and Table 4., Zelda was still found at high level of
binding. However, the transcription factor with
highest binding score at the 8656150bp peak is Trl as
shown in Table 3. Zen, bcd, and sna were also found
at high level around another Zelda binding peak at
8657916bp as shown in Table 4.
Analyzing Zelda and Other Transcription Factors That Regulating Ubiquitous and Patterning Genes of Drosophila Melanogaster
821
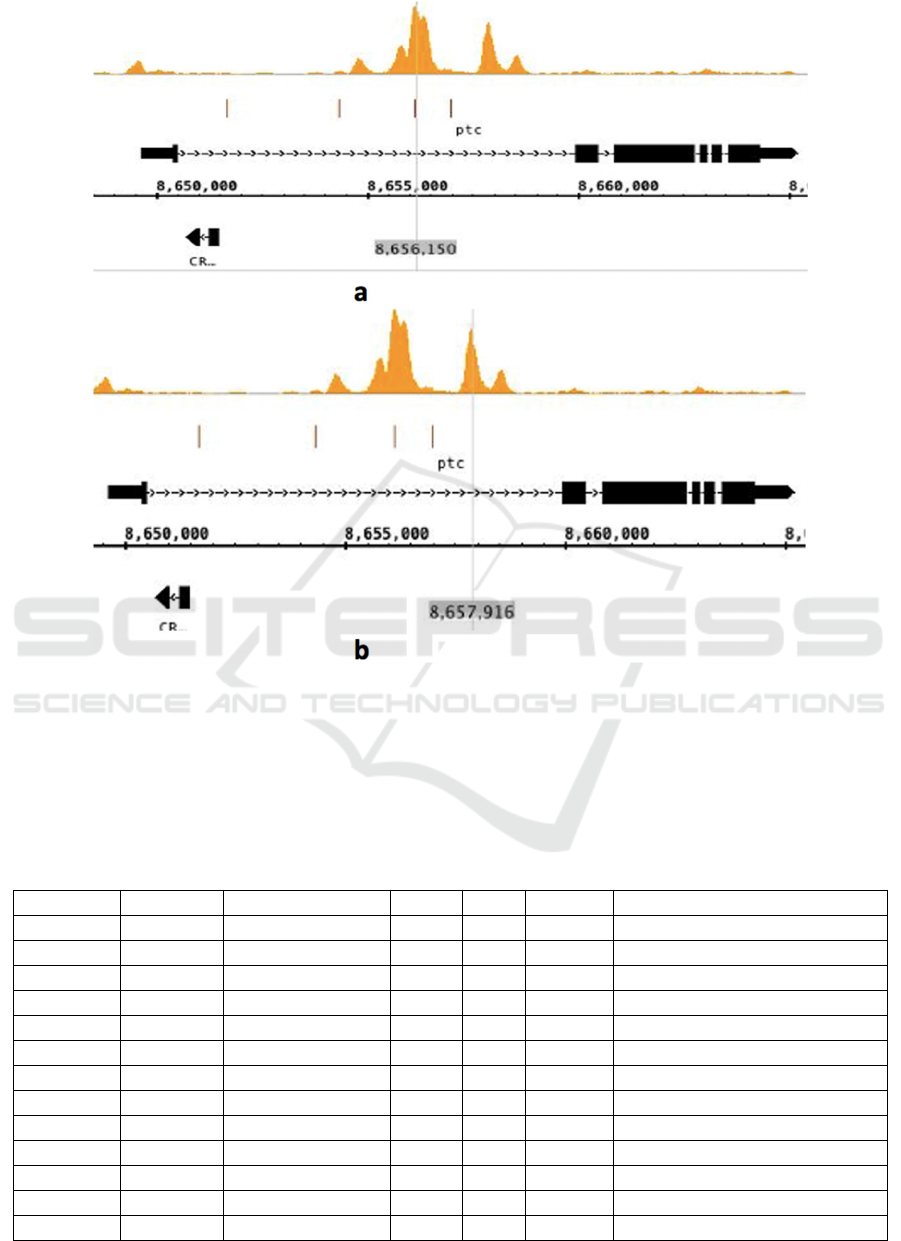
Figure 4: Zelda binding peaks at the enhancer region of Class 2 gene CG2411/ptc.
This figure incorporates 2 screenshots of the IGB
graph result of the Class 2 patterning gene
CG2411/ptc. 2 Zelda binding peaks are shown in a.
and b. respectively. The vertical grey lines show the
approximate Zelda binding peaks, together with grey
highlighted numbers representing the exact locations
of peaks in Chromosome 2R of Drosophila
melanogaster.
Table 3. Binding levels of different transcription factors around the Zelda binding peak 8656150 (Figure 4a.) in enhancer
region of CG2411/ptc.
Name Score Relative score Start End Strand Predicted sequence
Dsp1 15.9438 0.89694906 334 350 + CCAGAGAGAGAGGGAAG
Trl 15.0939 0.95595682 335 346 + CAGAGAGAGAGG
trx 13.4605 0.925088 336 347 + AGAGAGAGAGGG
Trl 13.234 0.92390239 337 348 + GAGAGAGAGGGA
trx 13.1356 0.9188802 338 349 + AGAGAGAGGGAA
vfl 12.2447 0.96162557 195 206 + GGCCAGGTAGGT
Trl 11.9072 0.90103495 333 344 + TCCAGAGAGAGA
Clamp 11.3008 0.83709098 335 348 + CAGAGAGAGAGGGA
exd 11.2783 1 320 327 + TTTTGACA
Clamp 11.2424 0.83616404 337 350 + GAGAGAGAGGGAAG
br
11.1682 0.88585636 230 243 + ATAATAAAGAAATT
Trl 11.0052 0.9572637 337 346 - CCTCTCTCTC
b
r(var.4) 11.0036 0.93649654 231 241 + TAATAAAGAAA
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
822
sna 10.8157 0.8877825 387 399 - GCGCCAGGTGCAA
vfl 10.754 0.93185675 181 192 - CCACAGGTACAC
sna 10.7444 1.00000002 275 280 + CAGGTG
sna 10.7444 1.00000002 390 395 - CAGGTG
b
r
k
10.5332 0.98996876 393 400 + CTGGCGCT
pn
r
10.4452 0.89460235 212 222 + TTTCGATTTTC
trx 10.4392 0.86735332 334 345 + CCAGAGAGAGAG
cad 10.4102 0.99753146 231 237 - TTTATTA
Stat92E 10.3244 0.85034053 39 53 + CGGAATTCACTGAAA
achi 10.2395 1 323 328 + TGACAG
Trl 10.1298 0.93081651 335 344 - TCTCTCTCTG
cad 10.0542 0.90889508 229 239 + AATAATAAAGA
Trl 10.0349 0.92794765 339 348 - TCCCTCTCTC
Table 4. Binding levels of different transcription factors around the Zelda binding peak 8657916 (Figure 4b.) in enhancer
region of CG2411/ptc.
Name Score Relative score Start End Strand Predicted sequence
vfl 13.955 0.99578006 105 116 - GTGCAGGTAGAC
sna 13.4007 0.93574936 111 123 - ACAGCAGGTGCAG
cad 12.3672 0.93962655 369 379 + GATCATAAATC
Ptx1 11.9592 0.99999999 238 244 - TTAATCC
dve 11.958 0.98124624 237 244 - TTAATCCG
CG11617 11.6102 1 19 25 - TTAACAT
nub 11.6076 0.86929265 18 29 + TATGTTAATCTG
od
d
11.5299 0.89574444 307 317 + AACAGCAGCAA
zen 11.0907 0.99999999 283 289 + CTAATGA
slp1 10.8339 0.91503434 130 140 + CTGTTTTCGTT
onecu
t
10.8156 0.99999999 375 381 - TTGATTT
sna 10.7444 1.00000002 114 119 - CAGGTG
b
cd 10.5191 1 238 243 - TAATCC
Sc
r
10.4198 0.96683999 283 289 + CTAATGA
oc 10.3923 1.00000001 238 243 - TAATCC
Abd-B 10.3839 1.00000001 371 377 - TTTATGA
so 10.3258 1.00000001 79 84 + TGATAC
in
d
10.3115 0.97777242 283 289 + CTAATGA
dl 10.2963 0.8678453 352 363 - CGGGGTTTCCTA
pb 10.2789 0.98339042 283 289 + CTAATGA
Gsc 10.1656 1 238 243 - TAATCC
nub 10.1448 0.83598283 34 45 + TATTTAAAATCG
3.4 Gene Expression Patterns
Figure 5. shows that Trithorax-like gene (Trl) and
Dorsal switch protein 1 (Dsp1) are expressed evenly
through the whole embryo. However, sna gene only
has perceptible level of expression in mesoderm at
the ventral side of the embryo.
Analyzing Zelda and Other Transcription Factors That Regulating Ubiquitous and Patterning Genes of Drosophila Melanogaster
823
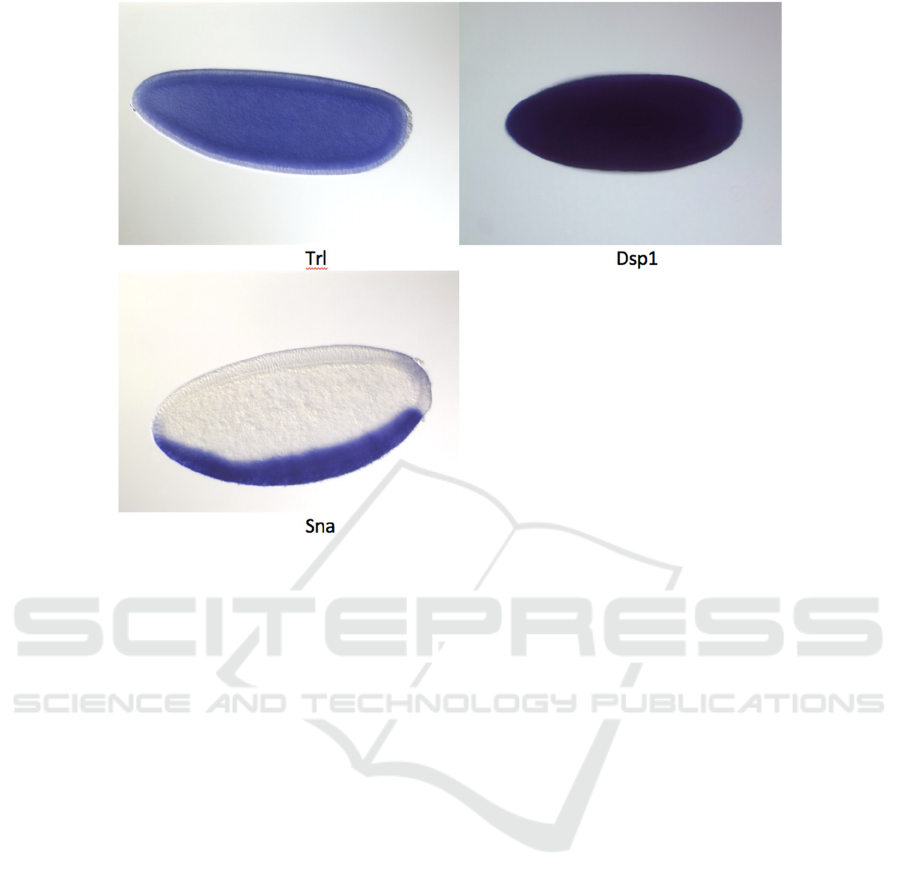
Figure 5. The gene expression results indicated by purple staining from Berkeley Drosophila Genome Project (BDGP).
The purple staining indicates the gene expression
during embryonic cycles 4 – 6. The darker the color,
the higher the expression level. The embryo shows
white instead of purple if there’s no expression. The
names of the genes are labelled below the images of
each gene.
4 DISCUSSIONS
In most cases, IGB graphs are reliable for us to
distinguish Class 1 and Class 2 genes. Comparing
graphs in Figure 1 with those in Figure 2, it is
obvious that the Class 1 gene transcription is
thoroughly turned off when Zelda is knocked out.
This is because Zelda binds to the promoter region of
the ubiquitous Class 1 gene to switch on the
transcription. That’s why removal of Zelda will
completely turn off the transcription of the gene and
make the RNA level reduced to zero. However, some
RNA molecules are still produced in CG4261/term
Figure 1. This is attributable to incomplete knockout
of Zelda. In comparison, Zelda binds to the enhancer
region of the patterning genes (CG2411/ptc and
CG4889/wg) Figure 2. In this case, Zelda works as
an activator and binds to the enhancer to stimulate the
transcription activity and produce more RNA
products. Therefore, the knockout of Zelda in
patterning genes will only reduce the amounts of
RNA transcribed instead of totally turned the
transcription off. Patterning gene promoters are
controlled by there own transcription factors. Another
obvious phenomenon in the IGB results is that Zelda
binding peaks are fused together at the promoter part
nearby the start of the Class 1 genes Figure 1.
However, in patterning genes Figure 2, Zelda peaks
are separate and they situate at the intron region of the
gene. This is another reliable property for us to
distinguish ubiquitous Class 1 genes from patterning
genes.
The JASPER result of CG1641/sisA in Table 1.
shows that Zelda (vfl) has the highest binding score
in the promoter region of sisA shown in Figure 3.
The binding sequence with the highest score contains
CAGGTAG, a common tag with high affinity to
Zelda. Zelda is an essential transcription factor for
early development of embryo (He et al 2019). It
enables the transcription of ubiquitous genes by
binding to the promoter and switching on the
transcription. That’s why Zelda can be found at such
high level here. Kruppel (Kr) is also found at high
binding score around the sisA gene’s Zelda binding
promoter region. This is a patterning gene that plays
an integral role in the segmentation of the embryo. In
contrast to ubiquitous genes, Kruppel’s expression is
uneven in the whole embryo. It is expressed in the
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
824

nervous and muscular system of the embryo and
regulates the thoracic and abdominal development.
Caudal (cad) is another patterning gene that is
responsible for anterior/posterior determination of the
embryo. Bicoid (bcd) is the transcription factor that
binds to the promoter of hunchback (hb), a patterning
gene expressed at head and tail of the embryo. Dorsal
(dl) is another patterning gene that has an expression
gradient that decreases as going from the ventral side
to the dorsal side. Which means dorsal expression is
highest at the mesoderm of the embryo’s ventral part
and close to zero at the embryo’s dorsal side.
Zerknullt (zen) expression can be inhibited by any
level of dorsal gene product, that’s why it’s a
patterning gene which can only be found at the dorsal
part of the embryo. CG1641/sisA is an ubiquitous
Class 1 gene whose expression is theoretically evenly
distributed through the whole embryo. However, it is
regulated by so many patterning genes that are
expressed at different parts of the embryo. This
further demonstrated that sisA can be found at
anywhere of the embryo. What’s more important is
that due to the regulation of so many patterning
transcription factors (Kr, cad, bcd, dl, zen), sisA
expression level is uneven across the whole embryo.
Therefore, Class 1 genes’ expression should be
everywhere in the embryo if the Zelda is functional
but the expression levels are diverse at different area
caused by the regulation of patterning gene products.
The JASPER result of the patterning gene,
CG2411/ptc, shown in Figure 4. and Table 3.4., also
indicates high level of Zelda binding. However, in
this case, Zelda binds to the enhancer of the gene to
increase the expression level instead of completely
controlling the switch on/off of the transcription.
Around the 8656150bp Zelda binding peak shown by
Figure 4a, there is also very high level binding of
Trithorax-like gene (Trl) (Table 3). The Trl gene
product is a transcription factor for chromatin
modification. Trl has even higher binding score to ptc
than Zelda. According to the expression patterning
shown in Figure 5, Trl is an ubiquitous Class 1 gene.
However the transcription factor with the highest
binding score here is Dorsal switch protein 1 (Dsp1)
according to Table 3. This transcription factor prefer
to bind a single strand of DNA molecules and
subsequently unwind the double-stranded DNA. It is
an ubiquitous gene according to the BDGP data
shown in Figure 5. It makes sense as all genes in the
embryo can only be transcribed after the double-
stranded DNA is unwound. CG2411/ptc has
patterning expression through the whole embryo, but
it is regulated by high level of ubiquitous gene
products such as Trl and Dsp1 situated at its enhancer
region. This demonstrated that Class 2 genes’
expression is also controlled by many Class 1 genes.
The reason might be that Class 1 genes have generic
functions required for transcription of all genes, the
unwinding function of Dsp1 is an example.
Therefore, no matter the gene’s expression is
patterning or ubiquitous, its transcription is regulated
by many ubiquitous Class 1 genes. The second Zelda
binding peak on CG2411/ptc is at 8657916bp and
shown by Figure 4b. In the 400bp region around this
peak, zen, bcd, and dl are also found at high level
according to Table 4. These three patterning genes
are mentioned above. Another transcription factor
found at high score is snail (sna), a patterning gene
that is essential for mesoderm development. Sna is
activated by high level dl and is expressed in
mesoderm at the ventral side of the embryo shown in
Figure 5. Sna is also found at high level around the
8656150bp Zelda binding peak represented by
Figure 4a. Although there’s no data of ptc’s
expression pattern in BDGP, we can estimate its
pattern according to the patterns of the patterning
gene which regulate its transcription. dl and sna are
expressed at mesoderm. Bicoid (bcd) is quite
important for head development and is expressed at
highest concentration in head. zen can only be found
at dorsal side of the embryo because of the inhibition
by dl. Therefore, we can estimate that ptc is expressed
mainly at the head, ventral and dorsal part of the
embryo.
5 CONCLUSIONS
In conclusion, graphs from IGB are reliable tools to
determine whether a Drosophila gene is Class 1
(ubiquitous) or Class 2 (patterning). According to
JASPAR data of the Class 1 CG1641/sisA and Class
2 CG2411/ptc, Class 1 gene is totally controlled by
Zelda that switch it on or off by directly binding to
the promoter. However, Zelda is not the only factor
controlling the Class 1 gene transcription. On the one
hand, there are many patterning gene transcription
factors that regulate the transcription level of the
Class 1 gene. That’s why the theoretically ubiquitous
expression level of Class 1 gene is actually uneven
across the embryo. On the other hand, Class 2 genes
are also regulated by ubiquitous Class 1 gene
products because some functions of these
ubiquitously expressed products can influence the
transcription rates of all genes in the organism. This
research, underpinned by powerful software and
pioneering databases, revealed the complexity of
genes’ network during early development of animals.
Analyzing Zelda and Other Transcription Factors That Regulating Ubiquitous and Patterning Genes of Drosophila Melanogaster
825

It also uncovered the essentiality of biotechnology
and modern data science to further researches in
genetics. All participated students were given
fascinating insight into the future of genetics.
REFERENCES
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K.,
& Walter, P. (2002). Drosophila and the Molecular
Genetics of Pattern Formation: Genesis of the Body
Plan. Molecular Biology of the Cell. 4th edition.
Garland Science, New York. Available from:
https://www.ncbi.nlm.nih.gov/books/NBK26906/
BDGP website https://insitu.fruitfly.org/cgi-
bin/ex/insitu.pl
He, S., Zhang, G., Wang, J., Gao, Y., Sun, R., Cao, Z.,
Chen, Z., Zheng, X., Yuan, J., Luo, Y., Wang, X.,
Zhang, W., Zhang, P., Zhao, Y., He, C., Tao, Y., Sun,
Q., & Chen, D. (2019). 6mA-DNA-binding factor Jumu
controls maternal-to-zygotic transition upstream of
Zelda. Nature communications, 10(1): 2219.
JASPAR website (2020) http://jaspar.genereg.net
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin,
Y., Albu, M., Chen, X., Taipale, J., Hughes, T. R., &
Weirauch, M. T. (2018). The Human Transcription
Factors. Cell, 172(4): 650–665.
Mikhaylichenko, O., Bondarenko, V., Harnett, D., Schor, I.
E., Males, M., Viales, R. R., & Furlong, E. (2018). The
degree of enhancer or promoter activity is reflected by
the levels and directionality of eRNA transcription.
Genes & development, 32(1): 42–57.
NCBI website https://www.ncbi.nlm.nih.gov
Signor, S. A., & Nuzhdin, S. V. (2018). The Evolution of
Gene Expression in cis and trans. Trends in genetics :
TIG, 34(7): 532–544.
Tolwinski N. S. (2017). Introduction: Drosophila-A Model
System for Developmental Biology. Journal of
developmental biology, 5(3): 9.
Ventos-Alfonso, A., Ylla, G., & Belles, X. (2019). Zelda
and the maternal-to-zygotic transition in cockroaches.
The FEBS journal, 286(16): 3206–3221.
Zabidi, M. A., & Stark, A. (2016). Regulatory Enhancer-
Core-Promoter Communication via Transcription
Factors and Cofactors. Trends in genetics : TIG,
32(12): 801–814.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
826
