
Large Scale Virtual Screening for Finding Inhibitor against the
RNA-dependent RNA Polymerase from Herbal Medicine for
SARS-Cov-2 Therapy
Xiaogang Liu
a
, Zirong Liang
b
, Shiye Wu
c
, Ying Wang
d
and Binquan Gou
*e
School of Materials and Environment, Beijing Institute of Technology, Zhuhai, 519000, China
Keywords:
SARS-Cov-2, Rdrp, TCM Database, Molecular Docking, Molecular Dynamics Simulation.
Abstract:
The COVID-19 pandemic caused by SARS-CoV-2 has cause worldwide health concerns. The research on
the virus infection mechanism was carried out and has made some progress. For the SARS-CoV-2
replication, the RNA-dependent RNA polymerase (RdRp) plays an important role and has been proven to be
an effective target. Traditional medicinal plants are widely used in epidemic prevention in China and they
have attracted great attention of scientists. Therefore, we executed large scale virtual screening on the
Traditional Chinese medicinal (TCM) database and hoped to find a potential drug against the virus
polymerase. As a result, we obtained nine non-toxic available compounds derived from TCM database by
docking and ADMET computing. Then 100 ns molecular dynamic was employed to uncover the potential
mechanisms which is helpful for further drug optimazation.
1 INTRODUCTION
1
A novel coronavirus that caused fatal respiratory
disease was reported at the end of 2019 (Zhu, Zhang,
Wang, Li, Yang, Song, Zhao, Huang, Shi, Lu, 2020).
The main clinical features of this rare virus disease
in the early stages were broadly fever, cough,
headache, diarrhea and loss of taste. As the infection
continues, some patients developed severe
respiratory difficulties, hemoptysis, diarrhea,
multiple-organ failure and death (Achak M, Alaoui
Bakri S, Chhiti Y, M'Hamdi Alaoui FE, Barka N,
Boumya W, 2021). The WHO experts warned that
virus can be spread quickly through close contact by
droplets or aerosols of cough and sneeze (Shereen
MA, Khan S, Kazmi A, Bashir N, Siddique R, 2020)
from an infected individual and has subsequently
named this pathogen 2019-nCoV (Ji, Wang, Zhao,
Zai, Li, 2020). Although government officials and
health experts around the world has implemented
measures to control its spread, the disease remains
a
https://orcid.org/0000-0002-5853-2005
b
https://orcid.org/0000-0002-4419-0999
c
https://orcid.org/0000-0001-5587-5304
d
https://orcid.org/0000-0002-7780-9372
e
https://orcid.org/0000-0003-2428-387X
great endemic worldwide. The 2019-nCoV led to
nearly 83 million infections and more than 1.8
million deaths (https://coronavirus.jhu.edu/).
Compared to the 1918 Spanish pandemic,
2019-nCoV spread had a disastrous impact on
human society (Shi, Wang, Shao, Huang, Gan,
Huang, Bucci E, Piacentini M, Ippolito G, Melino G,
2020).
With the help of whole genome sequencing
technology, Scientists have learned much of the
2019-nCoV and identified 2019-nCoV as a new
β-coronavirus (Wu, Zhao, Yu, Chen, Wang, Song,
Hu, Tao, Tian, Pei, 2020). As respiratory infectious
diseases SARS and MERS, the virus cause
respiratory infections in humans and scientists are
referring to it as SARS-CoV-2. The origin of this
virus is uncertain, but there is a definite
understanding of the lifecycle of the virus. The
diameter of the SARS-CoV-2 is 65-125 nm and
coated with glycoprotein on the outside and contains
26 to 32kbs positive-sense RNA inside the virus.
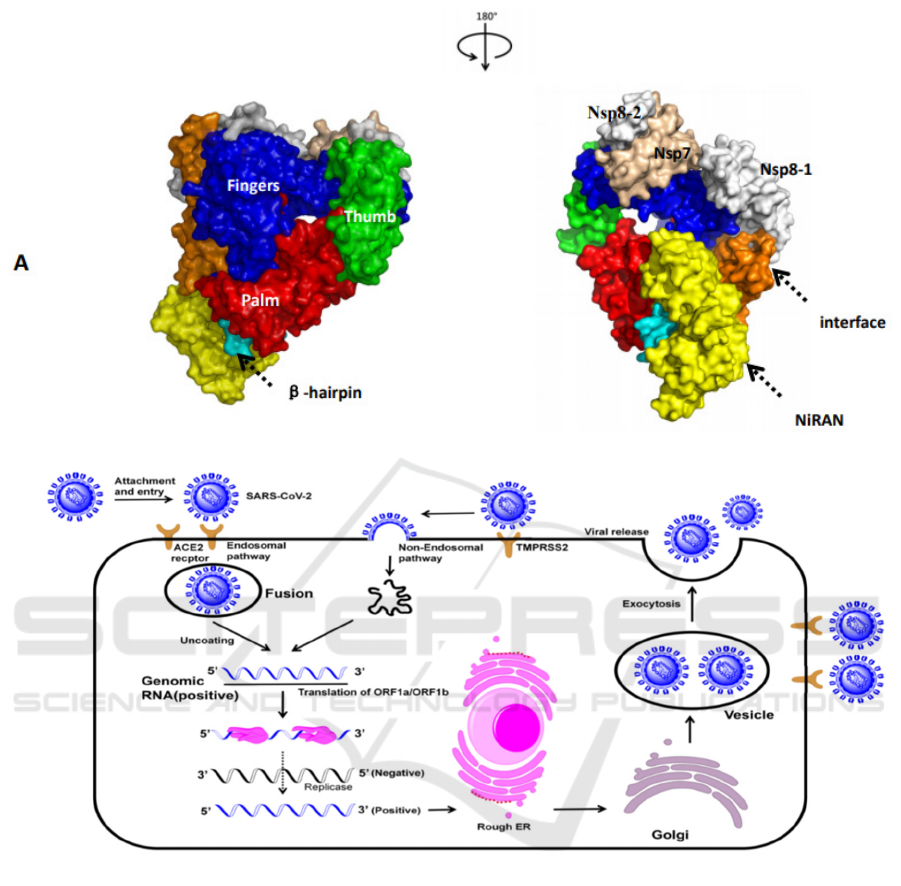
The SARS-CoV-2 infects cells through five typical
stages (Fig.1A). The first stage of the lifecycle is
attachment. With the spike glycoprotein which
recognizes the angiotensin converting enzyme 2 of
human cell, the virus completes the attachment and
enters cells by endocytosis (Luan, Lu, Jin, Zhang,
932
Liu, X., Liang, Z., Wu, S., Wang, Y. and Gou, B.
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2 Therapy.
DOI: 10.5220/0011313300003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 932-943
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2020). The second stage is membrane fusion. This
process is mediated by cathepsin-L in the endosome,
which promotes activation of spike protein and the
release of RNA. Alternatively, viruses fuse to cell
membranes by the membrane protein TMPRSS2 and
the genome enters the cell in a non-endocytic
pathway (Faheem, Kumar, Sekhar, Kunjiappan,
Jamalis, Balana-Fouce, Tekwani, Sankaranarayanan,
2020). Subsequently, the virus completes replication
and assembly in the host cell. The virus RNA
utilizes host cells ribosomes to synthesize two
polyproteins (pp1a and pp1ab) and four structural
proteins. The polyproteins are split into Nsp1-16
(non-structural proteins) by the papain-like domain
and the main protease contained in itself. Nsp3
contains a papain-like domain which slit
polyproteins at three different positions (Barretto,
Jukneliene, Ratia, Chen, Mesecar, Baker, 2005).
Meanwhile, Nsp5 is known as the main protease
which slit poly-proteins at eleven different positions
(Kumar, Bhardwaj, Kumar, Gehi, Kapuganti, Garg,
Nath, Giri, 2020). Nsp1 is associated with host
immune suppression (Kamitani, Huang, Narayanan,
Lokugamage, Makino, 2009) while the biological
function of the Nsp2 protein is unclear, and
scientists believe that the protein is associated with
the strong infectiousness of SARS (Angeletti,
Benvenuto, Bianchi, Giovanetti, Pascarella,
Ciccozzi, 2020). Nsp4, Nsp3 and Nsp6 are involved
in the formation of double-membrane vesicles
(Angelini, Akhlaghpour, Neuman, Buchmeier, 2013),
and Nsp7 and nsp8 act as co-factors to form
transcriptional complexes with the essential Nsp12,
which is directly involved in the replication of RNA
(Peng, Peng, Yuan, Zhao, Wang, Wang, Wang, Sun,
Fan, Qi, 2020). The Nsp11 is an intrinsically
disordered protein that may plays a critical role in
the interaction between virus and host cell
membrane (Gadhave, Kumar, Kumar, Bhardwaj,
Garg, Giri, 2021). The Nsp13 of the virus contains
three zinc ions and exhibits RNA helicase activity, it
plays a key role in the unwinding double- stranded
RNA when the virus replicate in host cells (Shu,
Huang, Wu, Ren, Zhang, Han, Mu, Wang, Qiu,
Zhang, 2020), which is similar to helicase
super-family-1.Nsp14 is a bifunctional enzyme
containing two structural domains, one for the
3’-5’exonuclease structural domain for RNA
proofreading and the other one is the transferase
active structural domain to produce cap structure of
7-methyl at the 5’-end of RNA guanine-N7 (Snijder,
Bredenbeek, Dobbe, Thiel, Ziebuhr, Poon, Guan,
Rozanov, Spaan, Gorbalenya, 2003). In contrast to
Nsp14, Nsp15 is a ribonucleic acid endonuclease.
Scientists believe that Nsp15 endonuclease activity
facilitates interference to the innate immune
response of the host (Kim, Jedrzejczak, Maltseva,
Wilamowski, Endres, Godzik, Michalska,
Joachimiak, 2020). Nsp16 plays an essential role in
immune evasion (Vithani, Ward, Zimmerman,
Novak, Borowsky, Singh, Bowman, 2020). With the
assistance of the non-structural proteins1-16, viral
replication and assembly are preforming in the
endoplasmic reticulum and Golgi apparatus of the
host cell. The last stage is exocytosis which Viral
particles are released outside the cell in a budding
(Mohanty, Sahoo, Padhy, 2021).
It is obvious that RdRp plays a critical role in the
replication and assembly of the SARS-CoV-2 in the
life cycle. Although the mechanism of RdRp is
unclear, we reconstructed the possible replication
patterns among the Non-structural proteins. It will
have positive implications for antiviral drug
development. The virus replication complex is
composed of three proteins: Nsp7, Nsp8 and Nsp12,
and the primers required in the virus replication
process are provided by Nsp7 and Nsp8 (Romano,
Ruggiero, Squeglia, Maga, Berisio, 2020). Nsp12 is
the critical enzyme that synthesizes viral dsRNA.
The RdRp complex contains six typical domains.
The function of the fingers and thumb domains is to
bind to the template chain. The palm structural
domain cooperates with the finger structural domain
to stabilize the phosphate skeleton, and catalyzes the
synthesis of RNA chains base on the principle of
base pairing (McDonald, 2013). Similar to RNA
family polymerases, the SARS-CoV-2 contains
seven conserved motifs in the palm structural
domain. The catalytic residues which are also
conserved in most viral RdRp is located in the motif
C (Gao, Huang, 2020). NiRAN domains are in close
contact with palm by an interface domain. β-hairpin
and NiRAN domaintogether stabilize the
polymerase structure.
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
933
B
Figure1: A: The structure of SARS-CoV-2 RdRp. B: The life cycle of SARS-CoV-2.
At present, effective drugs for the treatment of
SARS-Cov2 have not been reported. Some scientists
have found that the anti-virus RdRp drugs approved
by FDA are somewhat effective against
SARS-Cov-2. Ribavirin, Remdesivir, Sofosbuvir
and Galli- decivir (Elfiky, 2020) that target RNA
polymerase to inhibit viral replication are some of
the few drugs. But more experimental data are still
needed to support this discovery.
TCM is popular in the Chinese cultural circle.
This medicine based on life experience has been
practiced in East Asia for 5,000 years (Leng, Gany,
2014). TCM contains 11146 kinds of medicinal
plants and is a very valuable resource bank. There
have been numerous successful examples of finding
drugs from TCM. Computational technologies have
also made advances that can support these very large
scale screenings (Perez-Regidor, Zarioh, Ortega,
Martin-Santamaria, 2016). In this study the
SARS-CoV-2 RdRp is used as the template to search
for lead compounds from the TCM database. In
sight of the long history of traditional Chinese
medicine, so the drug candidates are also used in
emergency health events.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
934

2 METHODS
The calculation method of this study is illustrated in
Fig.2
2.1 Structural Analyses for the
SARS-Cov-2 RdRp
The crystal structure of SARS-CoV2 RdRp complex
(PDBID: 7AAP) was downloaded from the RCSB.
Protein primary structure and physicochemical
parameters of RdRp is performed using an online
server of ExPASy (Gasteiger, Gattiker, Hoogland,
Ivanyi, Appel, Bairoch, 2003)
.
MCPB. py can
perform parameterizations for both Zn
2+
ions
coordination bond (Li, Merz, Jr, 2016). The crystal
of 7AAP contains a gap between Leu895 and
Asn911.we employed Rosetta3.10 to build the gap
(Leman, Weitzner, Lewis, Adolf-Bryfogle, Alam,
Alford, Aprahamian, Baker, Barlow, Barth, 2020).
2.2 Molecular Docking
Traditional Chinese Medicine database contains
11146 kinds of medicinal plants and 33765
molecules. Molecular docking was carried out to
study the binding affinity between all molecules and
target protein (7AAP) by AutoDockVina1.2 (Trott O,
Olson AJ, 2010). Grid box coordinate was set at (x,
y, z) = 98.555, 96.343, 104.405 and docking
computing parameter employs AutoDock Vina
default settings. Pharmacokinetics assessment and
analysis is performed using an online ADMETsar
server (Cheng, Li, Zhou, Shen, Wu, Liu, Lee, Tang,
2012).
2.3 Molecular Dynamics Simulation
MD simulation computing was performed in explicit
solvent system. AMBER18 program was employed
to run 100ns for collecting data (Song, Lee, Zhu,
York, Merz, Jr, 2019). Force field of small
molecules, water and protein are generated
separately using antechamber, TIPT3P and ff14SB
of AMBER module (Maier, Martinez, Kasavajhala,
Wickstrom, Hauser, Simmerling, 2015; Steinberg,
Russo, Frey, 2019). The production simulation was
run 100 ns for three times.
2.4 RMSD and RMSF Calculation
Root mean squared deviation (RMSD) is used to
analyze the stability of complexes. When N means
the number of atoms, mi means the mass of atomi,
Xi means the coordinate vector for targetatom i, Yj
means the coor dinate vector for reference atom and
M means the total mass (Meli, Biggin, 2020; Khan,
Umbreen, Hameed, Fatima, Zahoor, Babar, Waseem,
Hussain, Rizwan, Zaman, 2021).
RMSD=
∑
m
i
∗X
i
−Y
j
2
N
i0
M
Root means square fluctuation (RMSF) analysis
was used to estimate the fluctuations of each amino
acid residue over the simulation time. When T is the
whole simulation time, tis the mass of atom i, ⎯ Xi is
the average coordinate for target residue i, x(t) is the
coordinate of residue i in time t.
𝑅𝑀𝑆𝐹=
1
𝑇
(𝑥
(
𝑡
)
−
𝑋
)
2.5 Binding Free Energy Calculation
The free energy of RdRp binding to small molecules
was calculated by the molecular mechanics energies
combined with the generalized born and surface area
continuum salvation (MM/PBSA) method (Jessica,
Swanson, Andrew McCammon, 2004). In order to
identify the most crucial residues of RdRp for the
binding of the natural small molecules, the total
binding free energy was decomposed into
contributions from individual residues (i = 1, 2, …,
932):
∆𝐺
=∆𝐺
=∆𝐺
,
∆G
bind
i
is the per-residue contributions, and
∆G
bind
i,j
isthe residue-pairwise interaction
contributions. The calculations were rendered by the
MMPBSA.py.MPI module of AMBER (Miller,
McGee, Jr., Swails, Homeyer, Gohlke, Roitberg,
2012).
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
935

Figure 2: Process of virtual screening.
Table1: Summary of top ranked Phytochemical screened against RdRp receptor docking score and binding free energy.
Phytochemical name
Plant source
Phytochemical
structure
Docking
Score
(kcal/mol)
Binding
free
Energy
(kcal/mol)
Residues interacting
with phytochemical*
7',8'-Dihydroxuxuarine Aα[39]
Maytenus chuchuhuasca
-12
-39.5029
±4.1410
Ala685, Ala688, Ile589,
Met601, Trp598, Ser592
Thr591, Gln815, Val588,
Lys593, Leu758, Cys813
Ala580, Lys577, Ile494,
Gln573, Arg569, Leu576
Gly 590
7,8-Dihydroisoxuxuarine Gα[39]
Maytenus chuchuhuasca
-11.8
-34.5054
±2.5765
Lys551, Ser549, Ala550,
Ile548, Hie439, Arg836
Cys813, Trp598, Lys593
Met601, Thr591, Ser759
Leu758, Ala547, Lys621
Asp618, Tyr619, Lys798
Pro620, Arg553
7,8-Dihydroisoxuxuarine Fα[39]
Maytenus chuchuhuasca
-11.7
-43.5704
±0.7284
Val560, Ala558, Lys500
Gly559, Thr565, Gly683,
Asn568, Ala685, Asp684,
Arg569, Gln573, Leu576,
Lys577, Ala580, Gly590,
Tyr689, Thr687, Ser682,
Leu544, Lys545, Gln408,
Asn543, Val557, Ser501,
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
936
7',8'-Dihydroxuxuarine Dβ[39]
Maytenus chuchuhuasca
-11.6
-34.4187
±1.1823
Gly590, Arg553, Lys545,
Ala547, Tyr546, Trp598
Asp865, Leu758, Cys813
Ser592, Lys593, Ile864
Phe594, Ser861, Thr591,
Ile589
Scabrans G5
[40]
Gentiana scabra
-11.6
-24.4734
±4.2354
Cys813, Ser759, Asp760,
Asp761, Ala547, Arg836,
His439, Lys551, Ser549,
Gln815, Ala550, Lys798,
Asp618, Glu811, Lys621,
Ile548, Asp623, Ser814,
Asp865, Leu862, Ile837,
Ala840, Pro832, Arg555
6",8'-Bisdiosquinone
[41]
Diospyros mafiensis
-11.3
-30.9596
±3.7960
Ala688, Leu576, Ala580,
Tyr689, Lys577, Val588
Gly590, Gln573, Ile589
Arg569, Asn496, Asn497,
Lys500, Cys813, Leu758,
Gln815, Lys593, Trp598,
Met601, Ser592, Phe812,
Thr591
Scabrans G4
[40]
Gentiana scabra Bunge
-11.3
-17.1623±2
.8491
Asp623, Arg624, Arg553,
Asp452, Ala554, Val557
Thr556, Asn691, Thr687,
Ser759, Lys545, Ile548
Ala547, Ser549, Ser814
Asp761, Asp618, Lys798
Pro620, Lys621, Tyr619
Cys622, Arg555, Asp760
Pusilatin C
[42]
Blasiaceae
-11.2
-20.7154
±2.2755
Arg566, Thr562, Lys497
Asp 681, Ala685, Gly680
Val554, Lys542, Ser811
Cys810, Asp758, Leu755
Asp 757, Ser756, Asn688
Ser679, Thr684, Tyr686
Leu573, Lys574, Gln570
Ile491, Val490, Val492,
Ala682
Isoxuxuarine Gβ
[39]
Maytenus chuchuhuasca
-11.0
-38.1405
±3.4054
Ala685, Asn496, Ala688,
Ile589, Ser759, Leu758
Ser814, Ile837, Arg836
Asp833, Asp865, Pro832
Gln815, Lys593, Cys813
Thr591, Gly590, Ala580
Tyr689, Lys577, Leu576
Gln573, Ile494, Arg569,
Val493, Val495, Val588
Galidesivir
[24]
Drugs used as control
-6.4
-25.8283
±1.7663
Ala685, Val560, Thr565,
Lys500, Ile562, Ser501,
Ala502, Ala512, Leu498
Tyr516, Asn497, Val495
Arg596, Gln573, Asn496
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
937
Table 2: Residues contributing large amount of negative (<-1.0KJ/mol)) energies towards RdRp.
ligand Residues
7',8'-Dihydroxuxuarine Aα
Ile589 Ala685 Ile494 Lys577 Gln573 Leu758
-1.9956 -1.9249 -1.3253 -1.2867 -1.2833 -1.2817
7,8-Dihydroisoxuxuarine
Fα
Leu576 Ala685 Arg569 Gly559 Lys577 Val560
-1.4921 -1.4501 -1.3727 -1.1525 -1.1460 -1.0501
Isoxuxuarine Gβ
Ser592 Thr591 Ile589 Ile494 Lys593 Cys813
-2.8722 -2.1811 -1.9397 -1.3814 -1.3792 -1.3575
Table 3: Hydrogen bond lifetime(H-bond) during the 100 ns MD simulation.
ligand Acceptor DonorH Donor Frac
7',8'-Dihydroxuxuarine Aα
ligand@O3 Gln_573@HE21 Gln_573@NE2 0.2240
ligand@O3 Gln_573@HE22 Gln_573@NE2 0.2205
7,8-Dihydroisoxuxuarine
Fα
ligand@O1 Tyr_689@HH Tyr_689@OH 0.3265
ligand@O2 Val_560@H Val_560@N 0.1920
Isoxuxuarine Gβ
Thr_591@O ligand@H1 ligand@O6 0.1930
ligand@O5 Gln_573@HE21 Gln_573@NE2 0.2775
A
B
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
938
C
D
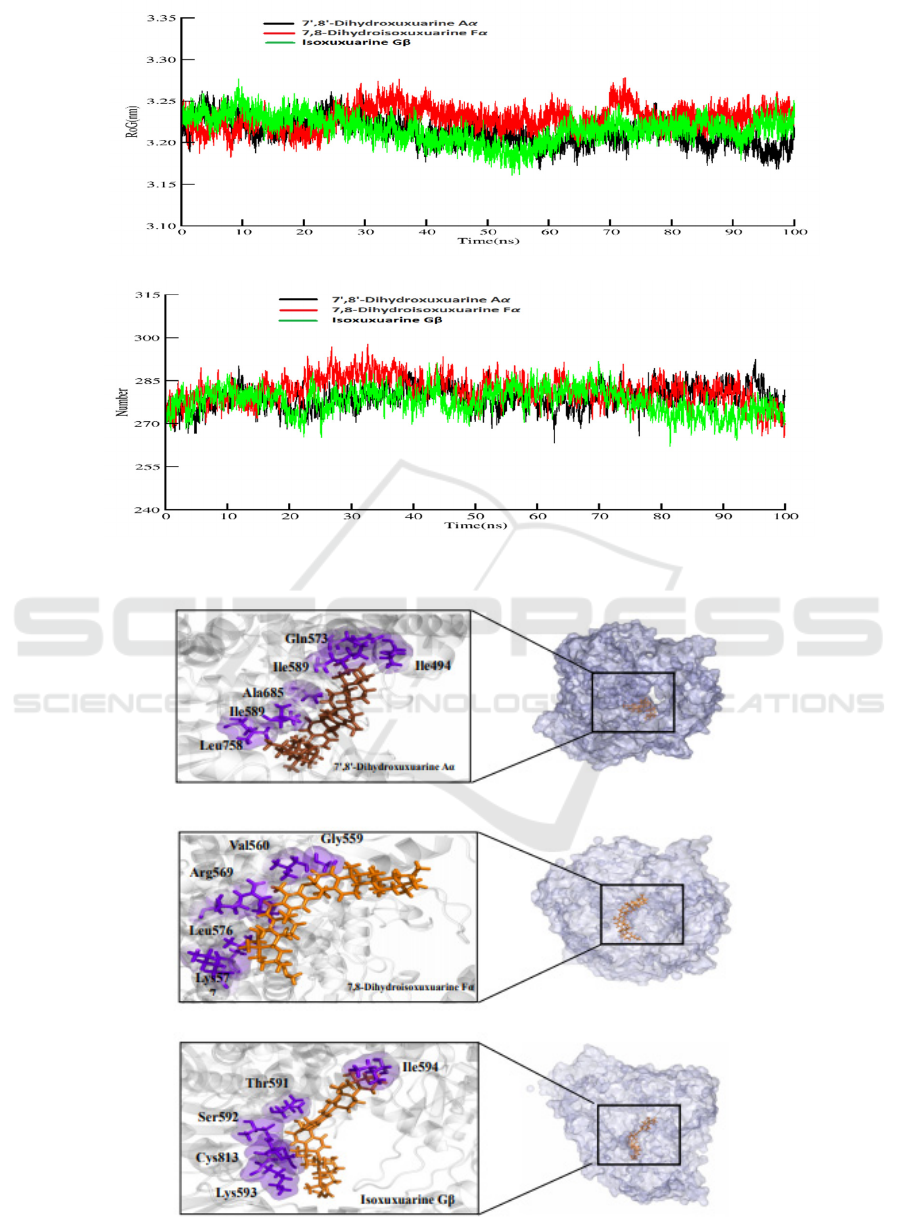
Figure 3: A: RMSD B: RMSF C: RoG D: H- bond interactions for three complexes in the 100ns simulation.
Figure 4: 3D structure for ligand-the SARS-CoV-2 RdRp.
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
939

3 RESULT
3.1 Structural of the SARS-CoV2
RdRp
TheSARS-CoV-2 RdRp contains 932 amino acids
and two zinc ion. One is bound to His295, Cys301,
Cys306 and Cys310 through four coordination
bonds. The other zinc ion is bound to His642,
Cys487, Cys645 and Cys646 through the four
coordination bonds. The molecular weight of the
SARS-CoV-2 RdRp is 106660.24 and the GRAVY
score is -0.224. Similar to RNA polymerase family,
the palm domain contains conversed motifs A-G.
The catalytic residue Asp760 of the SARS-CoV-2
RdRp is located in the motif C.
3.2 Molecular Docking
We docked Galidesivir, a potential inhibitor used as
control, with the binding groups and catalytic groups
of the SARS-CoV-2 Crystal RdRp Structure model
(PDBID: 7AAP). Traditional Chinese Medicine
databases which contain 33765 molecules were used
as the virtual screening protocol. Herein, we
identified small molecules that its docking scores is
higher than 11.0 kcal/mol by molecular docking
calculations. Then we screened 9 novel non-toxic
molecules thought the ADMETsar server (TableS1).
7',8'-Dihydroxuxuarine Aα was isolated from
Maytenus chuchuhuasca and exhibited the highest
docking score (-12kcal/mol). Isoxuxuarine Gβ was
also isolated from Maytenus chuchuhuasca and
exhibited the lowest docking score (-11.0kcal/mol).
Scabrans G5 was also isolated from Gentiana
scabra and forms hydrogen bonds with catalytic
residue Asp760 (Table 1; Fig S1), and exhibited the
middle docking score (-11.6kcal/mol).
3.3 RMSD and Free Energy
Decomposing
Amber18 was used to carry out 100ns MD
simulation for free energy calculation which result is
more accurate than that of docking (Table1 Binding
free energy). The production of MD simulation was
run for three times (Fig S2). This method can help
us understand what residues play important roles in
the complexes. From visualization of protein-ligand
system, and there are nine molecules locate at the
similar central cavity of the SARS-CoV-2 RdRp.
The three top of binding free energy is Isoxuxuarine
Gβ 7', 8'-Dihydroxuxuarine Aα, and 7,
8-Dihydroisoxuxuarine Fα. To do further
researching, we investigated the compactness,
stability and folding of protein through the Radius of
gyration calculation and fluctuations through
internal hydrogen bonds, two result indicates normal
behavior for three complexes (Figure3). Then we
also investigated the variation and stability of the
complexes through the RMSD calculation. As the
Figure3 shows, a similar trend of conformation
changes with RMSD over 932 C-alpha atoms for the
three systems during the whole simulation. It can be
found that the initial unsteady state lasted about
20ns before the atoms stably oscillated around their
new positions (0.3nm). Totally, a moderate
conformation change can be witnessed for the three
systems when compared them with their initial
structure.
Then we used RMSF to estimate the fluctuations
of each amino acid residue over the simulation time
(Figure 3). It is clear that different Phytochemical
cause fluctuation of different residues. For the
7',8'-Dihydroxuxuarine Aα-RdRP system, amino
acids fluctuate considerably around 16, 26, 61, 227,
263, 425, 595. However, the dramatic fluctuation
can be found around amino acids residues 62, 199,
227, 425, 853, 886 and 917 in the
7,8-Dihydroisoxuxuarine Fα-RdRp. Also, the
Isoxuxuarine Gβ mainly effects around residues
338,432, 644 and 824.
Based on the accurate binding free
energy(table2), In the 7',8'-Dihydroxuxuarine Aα
and Isoxuxuarine Gβ-RdRP, Ile589 and Ile494
contribute more to the free energy. On the contrary,
Ala685 and Lys577 contribute more to the binding
free energy in the 7',8'-Dihydroxuxuarine Aα and
7,8-Dihydroisoxuxuarine Fα
3.4 Hydrogen Bond Life Time Analysis
In this part, we focus on exploring the ligand and
protein interaction in details, which is helpful for
drug design and optimization in the future. H-Bond
plays an important role in structure-based drug
design. In general, the compounds will display great
activity if they interact with the key residues in the
protein. As the table 3 shows, the H atoms (HE21)
of Gln573 in the RdRp mainly interact O3 atom of
the 7', 8'-Dihydroxuxuarine Aα, accounting for 22%
in 100ns simulation. The H atoms (HH) of the
Tyr689 interact with O1 of the 7,
8-Dihydroisoxuxuarine Fα, accounting for 32%
within in 100 ns simulation. H atoms (HE21) of the
Gln573 interact with O5 Isoxuxuarine Gβ,
accounting for 27% within in 100 ns simulation.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
940

4 CONCLUSIONS
The growing SARS-CoV-2 cases urge the
development of specific drug and new therapeutics.
We investigated the life cycle of SARS-CoV-2 and
uncovered the mechanism of transmission. We
discovered that the SARS-CoV-2 RdRp plays a
unique role in viral replication and analyzed the
structure of the RdRp. It is obvious that the RdRp is
an ideal drug target. Considering the unique
contribution to epidemic prevention in the history,
traditional Chinese medicine database was screened.
Then we identified nine non-toxic compounds
though ADMET analysis. Finally, some detail
analysis was executed through the MD simulation,
such as the RMSD, RMSF, hydrogen bond lifetime,
free energy decomposing. The information may
contribute to the further drug design and
optimization. It is surprising that the top three
compounds come from Maytenus chuchuhuasca and
the plants are widely used in folk medicine in South
America (Osamu SHIROTA, 2004; Kikuchi,
Kakuda, Kikuchi, Yaoita, 2005; Ra®ullah M. Khana,
1999; Tatsuhiko Yoshida, Shigeru Takaoka, 1996;
Haydee Cha vez aGR, 2000). We anticipate that
the compounds screened from medical plants will be
used in the antiviral experiments and served as a
novel anti- SARS-CoV-2 drug.
FUNDING
This work was funded by Beijing Institute of
Technology, Zhuhai (Nos. XK-2019-03;
Nos.2020001TSZY)
CODE AVAILABLE
UCSD receipt # 2020-19-167
ACKNOWLEDGEMENTS
We thank National Supercomputing Center in
Shenzhen for providing the computational resources
and Gaussian09 software.
We also thank Beijing Computational Science
Research Center for providing Traditional Chinese
Medicine database
AUTHORS’ CONTRIBUTION
L.X and G.B performed the data processing; L.Z
advised on the implementation of image
decomposition and calculation of Euclidean
distances; all authors contributed to the final
manuscript
COMPETING INTERESTS
The authors have no competing interests.
REFERENCES
Achak M, Alaoui Bakri S, Chhiti Y, M'Hamdi Alaoui FE,
Barka N, Boumya W: SARS-CoV-2 in hospital
wastewater during outbreak of COVID-19: A review
on detection, survival and disinfection technologies.
Sci Total Environ 2021, 761:143192.
Angeletti S, Benvenuto D, Bianchi M, Giovanetti M,
Pascarella S, Ciccozzi M: COVID-2019: The role of
the nsp2 and nsp3 in its pathogenesis. J Med Virol
2020, 92(6):584-588.
Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier
MJ: Severe acute respiratory syndrome coronavirus
nonstructural proteins 3, 4, and 6 induce
double-membrane vesicles. mBio 2013, 4(4).
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD,
Baker SC: The papain-like protease of severe acute
respiratory syndrome coronavirus has deubiquitinating
activity. J Virol 2005, 79(24):15189-15198.
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW,
Tang Y: admetSAR: a comprehensive source and free
tool for assessment of chemical ADMET properties.
Journal of chemical information and modeling 2012,
52(11):3099-3105.
Elfiky AA: Ribavirin, Remdesivir, Sofosbuvir, Galidesivir,
and Tenofovir against SARS-CoV-2 RNA dependent
RNA polymerase (RdRp): A molecular docking study.
Life Sci 2020, 253:117592.
Faheem, Kumar BK, Sekhar K, Kunjiappan S, Jamalis J,
Balana-Fouce R, Tekwani BL, Sankaranarayanan M:
Druggable targets of SARS-CoV-2 and treatment
opportunities for COVID-19. Bioorg Chem 2020,
104:104269.
Gadhave K, Kumar P, Kumar A, Bhardwaj T, Garg N, Giri
R: Conformational dynamics of 13 amino acids long
NSP11 of SARS-CoV-2 under membrane mimetics
and different solvent conditions. Microb Pathog 2021,
158:105041.
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD,
Bairoch A: ExPASy: The proteomics server for
in-depth protein knowledge and analysis. Nucleic
Acids Res 2003, 31(13):3784-3788.
Haydee Cha vez aGR, a Ana Este vez-Braun, a,*:
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
941

Macrocarpins A±D, New Cytotoxic Nor-Triterpenes
from Maytenus macrocarpa. Bioorganic & Medicinal
Chemistry Letters 2000, 10 759±762.
Ji W, Wang W, Zhao X, Zai J, Li X: Cross-species
transmission of the newly identified coronavirus
2019-nCoV. J Med Virol 2020, 92(4):433-440.
Jessica M. J. Swanson RHH, and J. Andrew McCammon:
Revisiting Free Energy Calculations: A Theoretical
Connection to MM/PBSA and Direct Calculation of
the Association Free Energy. Biophysical Journal 2004,
86:67–74.
Kumar P, Bhardwaj T, Kumar A, Gehi BR, Kapuganti SK,
Garg N, Nath G, Giri R: Reprofiling of approved
drugs against SARS-CoV-2 main protease: an in-silico
study. J Biomol Struct Dyn 2020:1-15.
Kamitani W, Huang C, Narayanan K, Lokugamage KG,
Makino S: A two-pronged strategy to suppress host
protein synthesis by SARS coronavirus Nsp1 protein.
Nat Struct Mol Biol 2009, 16(11):1134-1140.
Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M,
Endres M, Godzik A, Michalska K, Joachimiak A:
Crystal structure of Nsp15 endoribonuclease NendoU
from SARS-CoV-2. Protein Sci 2020, 29 (7):
1596-1605.
Khan A, Umbreen S, Hameed A, Fatima R, Zahoor U,
Babar Z, Waseem M, Hussain Z, Rizwan M, Zaman N
et al: In Silico Mutagenesis-Based Remodelling of
SARS-CoV-1 Peptide (ATLQAIAS) to Inhibit
SARS-CoV-2: Structural-Dynamics and Free Energy
Calculations. Interdiscip Sci 2021, 13(3):521-534.
Kikuchi M, Kakuda R, Kikuchi M, Yaoita Y: Secoiridoid
glycosides from Gentiana scabra. J Nat Prod 2005,
68(5):751-753.
Luan J, Lu Y, Jin X, Zhang L: Spike protein recognition of
mammalian ACE2 predicts the host range and an
optimized ACE2 for SARS-CoV-2 infection. Biochem
Biophys Res Commun 2020, 526(1):165-169.
Li P, Merz KM, Jr.: MCPB.py: A Python Based Metal
Center Parameter Builder. Journal of chemical
information and modeling 2016, 56(4):599-604.
Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J,
Alam N, Alford RF, Aprahamian M, Baker D, Barlow
KA, Barth P et al: Macromolecular modeling and
design in Rosetta: recent methods and frameworks.
Nat Methods 2020, 17(7):665-680.
Leng JC, Gany F: Traditional Chinese medicine use
among Chinese immigrant cancer patients. J Cancer
Educ 2014, 29(1):56-61.
Mohanty SS, Sahoo CR, Padhy RN: Targeting Some
Enzymes with Repurposing Approved Pharmaceutical
Drugs for Expeditious Antiviral Approaches Against
Newer Strains of COVID-19. AAPS PharmSciTech
2021, 22(6):214.
McDonald SM: RNA synthetic mechanisms employed by
diverse families of RNA viruses. Wiley Interdiscip
Rev RNA 2013, 4(4):351-367.
Maier JA, Martinez C, Kasavajhala K, Wickstrom L,
Hauser KE, Simmerling C: ff14SB: Improving the
Accuracy of Protein Side Chain and Backbone
Parameters from ff99SB. J Chem Theory Comput
2015, 11(8):3696-3713.
Meli R, Biggin PC: spyrmsd: symmetry-corrected RMSD
calculations in Python. J Cheminform 2020, 12(1):49.
Miller BR, 3rd, McGee TD, Jr., Swails JM, Homeyer N,
Gohlke H, Roitberg AE: MMPBSA.py: An Efficient
Program for End-State Free Energy Calculations. J
Chem Theory Comput 2012, 8(9):3314-3321.
Osamu SHIROTA, a Setsuko SEKITA, a Motoyoshi
SATAKE, a Nine Regioisomeric and Stereoisomeric
Triterpene Dimers from Maytenus chuchuhuasca.
Chem Pharm Bull 2004, 52(6):739—746
Peng Q, Peng R, Yuan B, Zhao J, Wang M, Wang X, Wang
Q, Sun Y, Fan Z, Qi J et al: Structural and
Biochemical Characterization of the nsp12-nsp7-nsp8
Core Polymerase Complex from SARS-CoV-2. Cell
Rep 2020, 31(11):107774.
Perez-Regidor L, Zarioh M, Ortega L, Martin-Santamaria
S: Virtual Screening Approaches towards the
Discovery of Toll-Like Receptor Modulators. Int J
Mol Sci 2016, 17(9).
Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R:
A Structural View of SARS-CoV-2 RNA Replication
Machinery: RNA Synthesis, Proofreading and Final
Capping. Cells 2020, 9(5).
Ra®ullah M. Khana, Emil Rwekikab: 6'',
8''-Bisdiosquinone from Diospyros mafiensis.
Phytochemistry 1999, 50:143-146.
Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R:
COVID-19 infection: Origin, transmission, and
characteristics of human coronaviruses. J Adv Res
2020, 24:91-98.
Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci
E, Piacentini M, Ippolito G, Melino G: COVID-19
infection: the perspectives on immune responses. Cell
Death Differ 2020, 27(5):1451-1454.
Shu T, Huang M, Wu D, Ren Y, Zhang X, Han Y, Mu J,
Wang R, Qiu Y, Zhang DY et al: SARS-Coronavirus-2
Nsp13 Possesses NTPase and RNA Helicase Activities
That Can Be Inhibited by Bismuth Salts. Virol Sin
2020, 35(3):321-329.
Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J,
Poon LLM, Guan Y, Rozanov M, Spaan WJM,
Gorbalenya AE: Unique and Conserved Features of
Genome and Proteome of SARS-coronavirus, an Early
Split-off From the Coronavirus Group 2 Lineage.
Journal of Molecular Biology 2003, 331(5):991-1004.
Song LF, Lee TS, Zhu C, York DM, Merz KM, Jr.: Using
AMBER18 for Relative Free Energy Calculations. J
Chem Inf Model 2019, 59(7):3128-3135.
Steinberg L, Russo J, Frey J: A new topological descriptor
for water network structure. J Cheminform 2019,
11(1):48.
Trott O, Olson AJ: AutoDock Vina: improving the speed
and accuracy of docking with a new scoring function,
efficient optimization, and multithreading. J Comput
Chem 2010, 31(2):455-461.
Tatsuhiko Yoshida TH, Shigeru Takaoka: Phenolic
Constituents of the Liverwort: Four Novel Cyclic
Bisbibenzyl Dimers from Blasia pusilla L. 1996,
52(46):14487-14500.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
942

Vithani N, Ward MD, Zimmerman MI, Novak B,
Borowsky JH, Singh S, Bowman GR: SARS-CoV-2
Nsp16 activation mechanism and a cryptic pocket with
pan-coronavirus antiviral potential. bioRxiv 2020.
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y,
Tao ZW, Tian JH, Pei YY et al: A new coronavirus
associated with human respiratory disease in China.
Nature 2020, 579(7798):265-269.
Yan Gao LY, Yucen Huang: Structure of the
RNA-dependent RNA polymerase from COVID-19
virus. Science 2020, 368:779–782.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X,
Huang B, Shi W, Lu R et al: A Novel Coronavirus
from Patients with Pneumonia in China, 2019. N Engl
J Med 2020, 382(8):727-733.
Large Scale Virtual Screening for Finding Inhibitor against the RNA-dependent RNA Polymerase from Herbal Medicine for SARS-Cov-2
Therapy
943
