
Influence of Epigenetic Differences on the Etiology of Bipolar
Disorder and Schizophrenia
Zhiyu Yan
University College London, London, WC1H 0AJ, U.K.
Keywords: Epigenetic, Schizophrenia, Bipolar Disorder, Psychosis, DNA Methylation.
Abstract: Schizophrenia and bipolar disorder are two mental disorders that have attracted worldwide attention.
However, The role of DNA methylation in epigenetics in schizophrenia and bipolar disorder is unclear. This
paper investigates the role of epigenetics in the pathogenesis of schizophrenia and bipolar disorder.
Genome-wide analyses of monozygotic twins have identified specific genetic loci at which DNA
methylation may be responsible for schizophrenia and bipolar disorder. Certain conclusions were made by
genome-wide DNA methylation analysis of DNA samples from identical twins with inconsistent major
psychiatric disorders. In twins with schizophrenia, the largest differential methylation region associated with
mental illness was significant hypomethylation of the ST6GALNAC1 promoter, which overlaps with
previously reported rare schizophrenia genomes. The average difference in DNA methylation at this locus is
6%, but there is considerable variation between families, with some twins even showing a 20% difference in
methylation. These results suggest that DNA methylation differences play a role in phenotypic differences
in identical twins and may influence the etiology of schizophrenia and bipolar disorder to some extent.
1 INTRODUCTION
Schizophrenia and bipolar disorder are two related
mental disorders that are common across the globe
(Patel et al. 1996). Schizophrenia is a highly
inherited neuropsychiatric disease, which is mainly
manifested in the presence of psychotic symptoms,
but also characterized by dysfunctional emotional
response and cognitive changes. Although people
have succeeded in identifying the gene variants
associated with schizophrenia, they are still
uncertain about the pathogenic genes of the
pathogenesis of the disease and how their functions
are regulated (Hannon et al. 2016). Bipolar disorder
is an extremely debilitating mental illness. It is
characterized by paroxysmal mood swings. Patients
often experience both manic and depressive moods,
and often have cognitive impairment. People with
this disease have serious destructive attacks,
frequent recurrence and serious psychosocial
disorders. The disease usually begins in adolescence
and even in the late childhood of some patients,
much earlier than previously thought (Miklowitz et
al. 2008). Since the two diseases may have the same
etiology, the symptoms of schizophrenia and bipolar
disorder overlap and can be classified as major
psychosis (Cardno et al. 2002). Bipolar disorder and
schizophrenia have strong aggregation in the family.
Quantitative genetic analysis showed that both had
strong genetic components. However, although the
heritability of schizophrenia and bipolar disorder is
estimated to be 70%, the disease consistency of
monozygotic twins with the same DNA sequence is
not 100% (Cardno et al. 2000). This means that
non-genetic and environmental factors are also
important in the etiology of the diseases.
Epigenetics is a rapidly developing field that
includes regulatory mechanisms of gene expression
that do not involve genotype change. Epigenetic
mechanism mainly realizes heritable changes in gene
expression during mitosis through DNA methylation
and chromatin structure changes, but does not change
genomic DNA sequence. And the study of
epigenetics is increasingly relevant to neuroscience.
Epigenetic mechanisms involve brain development
and neuronal differentiation. Epigenetic regulation
involves multiple levels of gene expression, with
direct modifications from DNA and histone tails that
regulate transcription levels to interactions with
messenger RNA that regulate translation levels (Roy
et al. 2015). It is generally believed that epigenetic
dysfunction of human brain can be related to a series
Yan, Z.
Influence of Epigenetic Differences on the Etiology of Bipolar Disorder and Schizophrenia.
DOI: 10.5220/0011372300003438
In Proceedings of the 1st International Conference on Health Big Data and Intelligent Healthcare (ICHIH 2022), pages 471-477
ISBN: 978-989-758-596-8
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
471

of mental diseases, including psychosis. In recent
years, researchers have studied the brains of
psychiatric patients and healthy controls, and found
that there are significant epigenetic changes in the
genome related to schizophrenia and bipolar disorder
(Mill et al. 2008). But how the epigenome plays a
role in schizophrenia and bipolar disorder is not well
understood. The role of DNA methylation in
epigenetics in schizophrenia and bipolar disorder is
unclear.
Monozygotic twins carrying the same disease
mutation can be clinically quite different, and
investigating inconsistent monozygotic twins pairs is
a useful method for discovering disease-related
epigenetic mechanisms, as It can detect the
epigenome independently of potential variation of
genome sequence (Bell et al. 20011. A study found
substantial differences in DNA methylation variation
between monozygotic twins, suggesting that
epigenetic variation can lead to phenotypic
inconsistencies between humans with the same gene
(Kaminsky et al. 2009).
Because the role of DNA methylation in
schizophrenia and bipolar disorder is unclear, it is not
known exactly which DNA sites are involved. The
new study proposes genome-wide analysis to identify
the genetic loci most affected by the two diseases.
This paper investigates the role of epigenetics in
the pathogenesis of schizophrenia and bipolar
disorder. Genome-wide analyses of monozygotic
twins have identified specific genetic loci at which
DNA methylation may be responsible for
schizophrenia and bipolar disorder. Genome-wide
analysis of DNA methylation variations in identical
twins caused by schizophrenia and bipolar disorder,
with genetically damaged DNA extracted from a
unique twin. Many DNA methylation differences
associated with disease were found, many of which
were located near genes previously associated with
psychosis. The results agree with the hypothesis that
epigenetic changes can influence the causes of
schizophrenia and bipolar disorder (Emma et al.
2011).
Table 1: Group of monozygotic twin pairs utilised in the study, values shown are average plus standard deviation.
Schizophrenia-discordant twin
p
airs
Bipolar Disorder-discordant twin
p
airs
Psychosis-discordant twin
p
airs
Sex
(
males:females
)
8:3 2:9 10:12
Ethnicity 10 Caucasian, 1 unknown 10 Caucasian, 1 Afro-Caribbean
20 Caucasian, 1 unknown, 1
Afro-Caribbean
Time discordant
(years)
10.4 ± 10.6 14.6 ± 10.7 12.6 ± 10.6
Age of onset (years) 20.0 ± 4.6 21.7 ± 12.3 20.9 ± 9.3
2 METHODOLOGY
Since monozygotic twins share the same genetic
sequence, studying epigenetic changes in diseases in
inconsistent identical twins is a powerful approach
because it allows independent epigenetic assessment
of any potential genome sequence variation.
Inconsistent DNA methylation in schizophrenia
or bipolar disorder was measured in 22 pairs of
twins (44 individuals) using the Illumina Infinium
HumanMethylation27 BeadChip. Standard protocols
is used to extract genomic DNA from whole blood
of 22 pairs of inconsistent monozygotic twins
recorded in the Maudsley Bipolar disorder and
Schizophrenia Twin Study. In the experiment, the
twins were clinically diagnosed by at least two
psychiatrists and two psychologists to ensure mental
inconsistencies. On average, the twins had been ill
for 12.6 (+ 10.6) years when they were taken blood
(Table 1). The EZ 96-DNA methylation Kit (Zymo
Research, CA, USA) is used to replicate 500 nm of
genomic DNA per person with sodium bisulfite. In
order to study genome-wide DNA methylation,
Illumina Infinium human methylation 27 beadchip
was mainly used. The chip investigated 27578 CpG
sites related to about 14000 genes. Also, Illumina
GenomeStudio software play a important role in
extracting the signal strength of each probe and
perform an quality control inspection when all data
sets are available. The probes with p value of 0.05 (n
= 733) detected in all samples were removed, and
the probes with poor quality were strictly controlled.
This experiment mainly uses microarray data
analysis. In the experimental analysis procedures, all
calculations and statistical analyses are performed in
the R statistical analysis environment. The ratio of
the normalized signals of methylated probes to the
sum of the normalized signals of methylated and
unmethylated probes calculates the relative
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
472
methylation levels of each detected CpG site. This
gives an mean β value for each CpG loci, which is
from 0 to 1, where 0 means unmethylated and 1
means fully methylated.
In genome-wide correlation analysis, variable
probes are recorded by calculating the standard
deviation of the entire data set, and then those
probes whose standard deviation is less than the
estimated standard deviation are filtered out. Two
independent ranking tests were analyzed. The first is
the standard paired T-test, which looks at the
meaning of differences in DNA methylation
between the members of each twin pair. The another
test was used to measure the size of the methylation
difference, with a calculated Δβ-value describing the
average difference in methylation between the
members of each twin pair (Δβ refers to the
unaffected minus the affected twin). The results of
the two tests were sorted by P value and size. Next,
the two ranking lists were added together to produce
the final CpG site ranking list. The table shows the
CpG sites with the greatest differences in DNA
methylation and the most consistent in all twins. For
statistical analysis of the magnitude of change
observed at each locus for affected and unaffected
twins, the custom weighted T-test was used.
3 RESULTS
DNA methylation at a single CpG site illustrated
important difference in monozygotic twins.
Analytical methods was used to determine the
biggest differences in DNA methylation at specific
CpG sites. Many sites in the whole genome is
identified which showed differences in
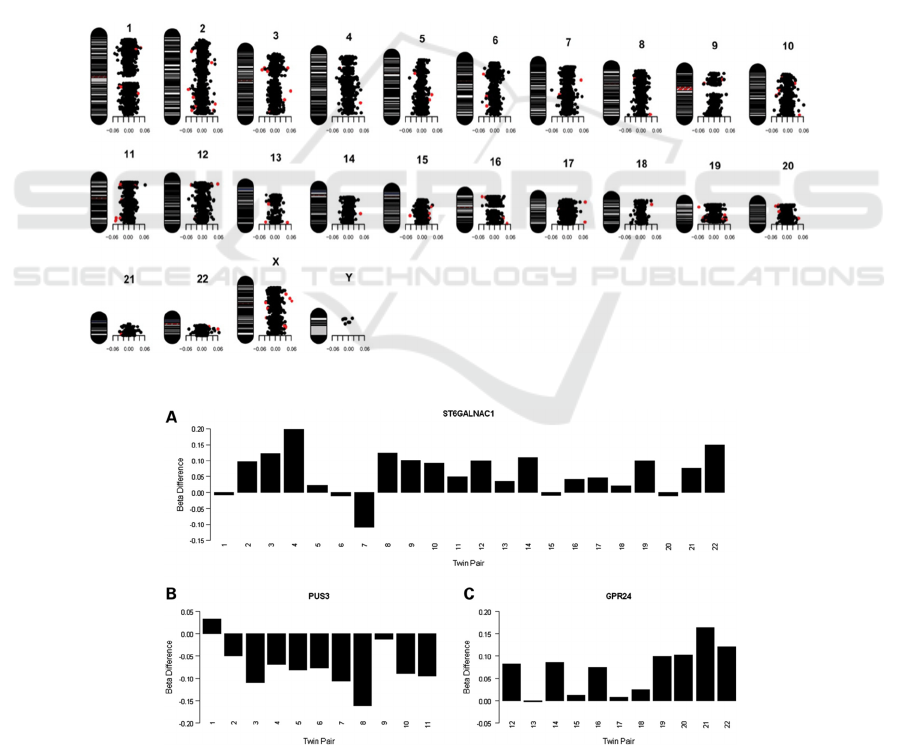
disease-related DNA methylation (Figures 1 & 2).
Figure 1: Characteristic map of each CpG locus in all 44 individuals of psychotic discordant twins.
Figure 2: DNA methylation differences (Δβ-value) for the top-ranked probes from (A) the combined psychosis-discordant
analysis group: ST6GALNAC1 (cg13015534), (B) Schizophrenia-discordant analysis group: PUS3 (cg02659232) and (C)
the bipolar disorder-discordant analysis group: GPR24 (cg21342728).
Influence of Epigenetic Differences on the Etiology of Bipolar Disorder and Schizophrenia
473
Table 2 shows the genomes closest to the
differentially methylated CpG sites associated with
the eight top diseases in the three experimental
groups (Schizophrenia, bipolar disorder and
combined psychosis). By analyzing the location of
100 CpG sites with the highest methylation
difference associated with psychosis, the
representation of CpG sites in CpG island was
significantly insufficient.
Table 2: The first eight of the three test groups had differential methylation CpG sites.
Analysis
g
rou
p
Rank Gene name Chromosome Paired t-test P-value
Mean Δβ (minimum–
maximum
)
Weighted Δβ
P-value
Weighted q-
value
Psychosis
1
ST6GALNA
C1
17q25.1 4.03E − 04 0.06 (−0.11–0.20) 1.19E − 07 7.97E − 04
2 ACADL 2
q
34 2.49E − 04 0.05
(
−0.05
–
0.17
)
1.59E − 06 3.19E − 03
3 TBC1D10A 22
q
12.2 8.56E − 04 0.06
(
−0.05
–
0.23
)
9.40E − 08 7.97E − 04
4 PUS3 11q24.2 7.66E − 04 −0.05 (−0.16
–
0.06) 1.71E − 06 3.19E − 03
5 FXR2 17p13.1 1.74E − 03 0.06 (−0.09
–
0.26) 9.03E − 08 7.97E − 04
6 TSP50 3p21.31 4.92E − 04 0.04 (−0.05
–
0.12) 2.26E − 05 1.12E − 02
7 PCOLN3 16
q
24.3 1.27E − 03 0.04
(
−0.06
–
0.21
)
1.79E − 05 1.03E − 02
8 SOX1 13
q
34 1.04E − 03 0.04
(
−0.04
–
0.13
)
2.64E − 05 1.18E − 02
Schizophr
enia
1 PUS3 11
q
24.2 7.66E − 04 −0.07
(
−0.16
–
0.03
)
5.16E − 05 0.10
2 SYNGR2 17q25.3 8.29E − 04 0.07 (0.01
–
0.13) 9.82E − 05 0.14
3 KDELR1 19q13.3 1.25E − 03 −0.06 (−0.14
–
0.01) 3.07E − 04 0.18
4 PDK3 Xp22.11 7.54E − 04 0.06 (0.00
–
0.14) 3.67E − 04 0.18
5 PPARGC1A 4
p
15.1 1.85E − 03 0.06
(
−0.02
–
0.12
)
2.89E − 04 0.18
6 ACADL 2
q
34 3.74E − 03 0.07
(
0.00
–
0.17
)
7.81E − 05 0.12
7 FLJ90650 5q23.1 4.19E − 04 0.05 (−0.01
–
0.09) 6.98E − 04 0.19
8 TUBB6 18p11.21 3.54E − 03 0.06 (−0.01
–
0.18) 1.78E − 04 0.16
Bipolar
disorder
1 GPR24 22q13.2 1.30E − 03 0.07 (0.00
–
0.16) 7.59E − 05 0.17
2 TLE6 19
p
13.3 1.97E − 03 −0.09
(
−0.21
–
0.01
)
1.54E − 05 0.12
3 STAB1 3
p
21.1 1.63E − 03 −0.07
(
−0.18
–
0.02
)
8.11E − 05 0.17
4 PPYR1 10
q
11.2 5.13E − 04 −0.06
(
−0.12
–
0.01
)
3.44E − 04 0.25
5 CTNNA2 2p12 3.59E − 03 0.09 (0.00
–
0.21) 1.56E − 05 0.12
6
ST6GALNA
C1
17q25.1 3.06E − 03 0.06 (−0.01–0.15) 2.82E − 04 0.23
7 C1orf35 1
q
42.13 5.30E − 03 0.06
(
−0.01
–
0.18
)
1.88E − 04 0.23
8 IQCH 15q23 3.94E − 03 0.05 (−0.06
–
0.10) 7.81E − 04 0.30
In all 22 pairs of uncoordinated monozygotic
twins, the methylation difference in the promoter site
of the gene sequence ST6GALNAC1 was the
highest, and the methylation degree of the affected
individuals was lower than that of the unaffected
monozygotic twins. The highest differentially
methylated CpG locus in discordant schizophrenic
twins is located upstream of the gene encoding
PUS3, which is highly methylated in affected twins.
The higher ranked CpG locus in inconsistent
comorbid psychiatric pairs is located upwards of the
gene site GPR24, which is methylated in diseased
twins.
Although the influence of the top loci in each
diagnostic group was in the same direction, the
significance of methylation difference between the
diseased and non-diseased twins at some loci was
different, suggesting familial heterogeneity even at
the top loci. The first psychotic differential
methylation site in the ST6GALNAC1 promoter
showed binomial Δβ values as high as 0.20 (Table
2). Psychosis has significant clinical heterogeneity,
so it is generally accepted that there are some rare
etiologies that are highly influential in some cases.
Individual twins may also vary greatly in
methylation at sites associated with particular
diseases (Merikangas et al. 2009). Therefore, DNA
methylation differences in individual twins to find
the greatest specific changes involved In families.
Many new and known psychiatric candidate genes
show significant differences in DNA methylation
between patients of one or more pairs of twins, and
some sites can be calculated with Δβ values greater
than 0.60 between diseased and non-diseased twins.
Some CpG loci ranked higher in schizophrenic
discordant and bipolar disorder discordant twin
analyses, but DNA methylation changes were
reversed between diseases, suggesting that
epigenetic mechanisms can be disease identified. In
monozygotic twins with dissonant schizophrenia and
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
474
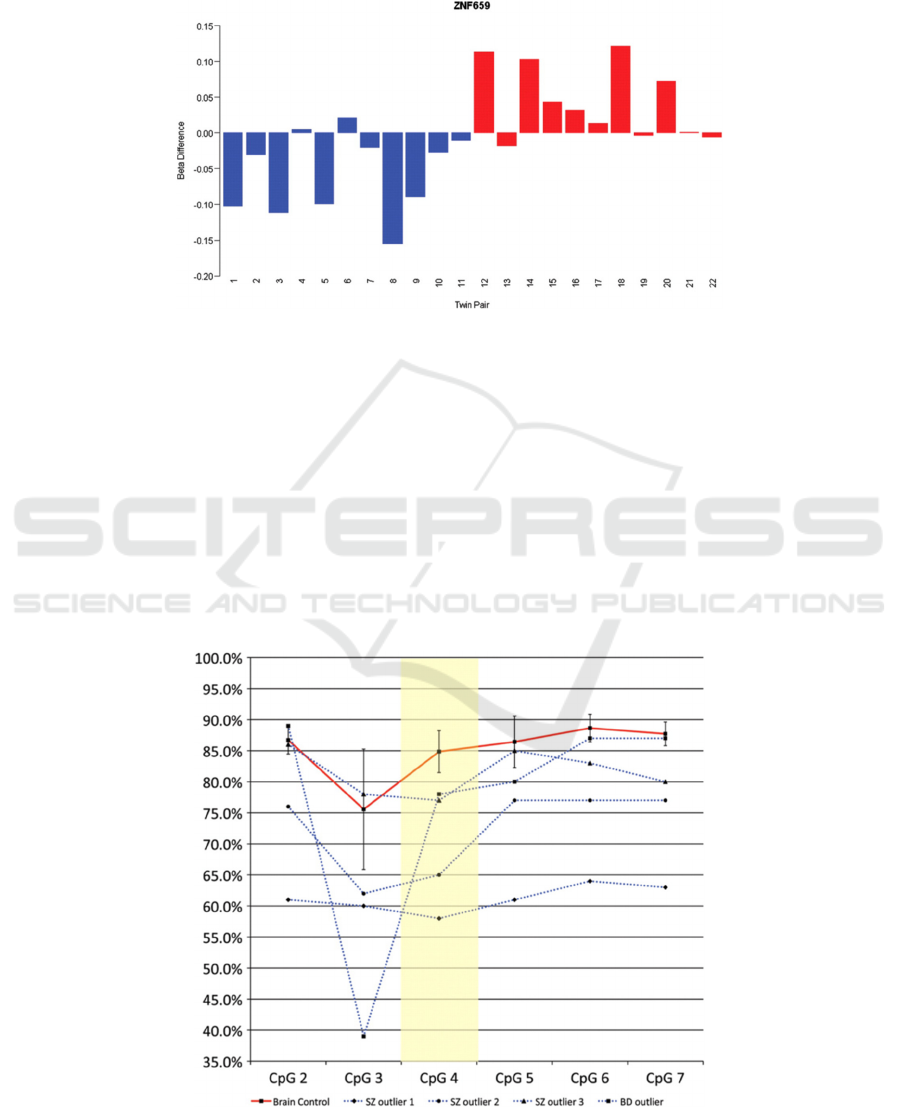
bipolar disorder, the CpG loci in the ZNF659
promoter was the highest of 100 sites and was
hypomethylated in bipolar disorder twins and
hypermethylated in schizophrenia twins (Figure 3).
Among the 100 highest-ranked schizophrenia and
bipolar disorder loci, several other studies have
shown commonly large diversities in the alternative
diagnostic group in the opposite direction.
Figure 3: DNA methylation differences (Δβ-value) for a CpG loci located at ZNF659 (cg18267381), where Schizophrenia
group is blue bars, and Bipolar disorder group is red bars.
Investigating that the validation and reproduction
of disease-associated unmethylated at the
ST6GALNAC1 gene loci. The Sequenom
EpiTYPER data, which accurately repeated arrays of
specific CpG sites on the Illumina platform, showed
hypommethylation in diseased psychiatric twins
(mean ¼ 35% methylation in psychotic twins and ¼
41% methylation in unaffected twins). Notably,
validation data for schizophrenic twins showed
greater differences in DNA methylation (an average
of 15% hypomethylation) between the diseased and
undiseased twins than was spotted at this locus. The
overall level of DNA methylation in the brain (85%)
was higher than that in the blood (40%). Although
no overall significant difference was found between
psychiatric patients and the control group at the
dominant site of DNA methylation array (CpG4
sequenom analysis), thirty (13.3%) psychiatric
patients tested illustrated clear (27%) methylated
and large difference at this CpG and many similar
CpG loci (Fig. 4).
Figure 4: DNA methylation at several CpG sites in the ST6GALNAC1 promoter of psychotic patients was compared with
postmortem brain tissue of controls.
Influence of Epigenetic Differences on the Etiology of Bipolar Disorder and Schizophrenia
475

This hypomethylated ST6GALNAC1 promoter
was not found in any of the control groups in this
study, indicating consistent and consistent
methylation in the area. These brain data flesh back
large heterogeneity in blood data between twins
(Figure 1), with some affected siblings showing
hypomethylation of up to 20%, but other twins
showing smaller differences. These data suggest that
major changes associated with disease in this site
may affect some patients.
4 DISCUSSION
A genome-wide approach was used to
comprehensively analyze disease-related DNA
methylation differences in monozygotic twins with
discorrelating schizophrenia and bipolar disorder. It
is preliminarily found that no significant changes in
total DNA methylation between the diseased and
undisased twins, but there were important
disease-related changes between the twins at
identified sites on the genome. Some methylation
differences persisted in the comorbidities, while
other differences may be specific to schizophrenia or
bipolar disorder. Although large differences were
found in nearly all pairs of discordant twins in each
diagnostic category, other differences related to
specificity for schizophrenia or bipolar disorder
were found for only one or a few pairs of twins.
Although these sites in the experiment had not
previously been linked to mental illness, evidence of
DNA methylation differences in genes involved in
mental illness could still be found. This illustrates
the data of this study and provides more
demonstrations to support the usage of DNA
methylation in the etiology of schizophrenia and
bipolar disorder.
The first total difference in methylation
combined with mental illness found in the study was
a CpG loci (17q25.1) in the ST6GALNAC1
promoter, and it is hypomethylation at this site in the
ill twins. They also found that 0.13% of autopsy
brain samples from patients with schizophrenia and
bipolar disorder showed significant hypomethylation
in extended regions containing the designated CpG
locus, which could indicate that epigenetic
mechanisms in this area can exist in a subset of
patients with psychosis. CpG sites associated with
mental illness do not exist in the same CpG island,
and differential methylation sites are significantly
deficient in classical CpG-rich promoters,
suggesting that phenotypic related variations in
DNA methylation generally occur outside these
regions, which is consistent with data from another
epigenomic analysis study (Weber et al. 2007).
However, the study did have some limitations.
For example, it was limited to 22 monozygotic
twins, calculations are based on standard deviation
estimates of the entire dataset from Illumina's array
data showing that at a strict Bonferroni-corrected
assumed value cutoff. The 22 monozygotic twins in
the study provided greater than 80% accuracy in
obtaining Δβ=0.06, although the ability to find small
differences at this level of significance was limited.
5 CONCLUSIONS
The role of epigenetics in the etiology of
schizophrenia and bipolar disorder has been
demonstrated experimentally. Genome-wide
analysis of monozygotic twins found that specific
sites of DNA methylation also contribute to the
cause of schizophrenia and bipolar disorder. In
patients with schizophrenia, the largest differential
methylation region associated with psychiatric
illness was hypomethylation of the ST6GALNAC1
promoter. The average difference in DNA
methylation at this site is 6%. However, there is
significant heterogeneity between families, since
some twin pairs having as much as 20% difference
in methylation. These results suggest that DNA
methylation differences play a role in phenotypic
differences in identical twins and may influence the
etiology of schizophrenia and bipolar disorder to
some extent.
This study provides insight into the etiology of
mental illness, particularly schizophrenia and
bipolar disorder. However, it is not particularly clear
how DNA methylation at specific sites affects
patients with both diseases. Epigenetics with
neuroscience is a hot research direction in recent
years, and great progress has been made in the
genetic etiology and treatment of mental diseases. In
the future, human diseases can be better understood
at the epigenome level by studying different
epigenetic modifications of DNA.
ACKNOWLEDGEMENTS
I would like to thank Prof. Wang and the supervisor
Ms. Wang for their guidance and academic
explanation of my paper.
ICHIH 2022 - International Conference on Health Big Data and Intelligent Healthcare
476

REFERENCES
Bell, J.T. and Spector, T. D. (2011). A twin approach to
unraveling epigenetics. Trends Genet., 27, pp.116–
125.
Cardno, A.G. and Gottesman, I. I. (2000). Twin studies of
schizophrenia:from bow-and-arrow concordances to
star wars Mx and functional genomics. Am. J. Med.
Genet., pp.97, 12–17.
Cardno, A.G., Rijsdijk, F.V., Sham, P.C., Murray, R.M.
and McGuffin, P. (2002). A twin study of genetic
relationships between psychotic symptoms. Am. J.
Psychiatry, 159, pp.539–545.
Emma L. Dempster, Ruth Pidsley, Leonard C. Schalkwyk,
Sheena Owens, Anna Georgiades, Fergus Kane,
Sridevi Kalidindi, Marco Picchioni, Eugenia Kravariti,
Timothea Toulopoulou, Robin M. Murray, Jonathan
Mill. (2011). Disease-associated epigenetic changes in
monozygotic twins discordant for schizophrenia and
bipolar disorder, Human Molecular Genetics, Volume
20, Issue 24, pp. 4786–4796.
Hannon, E., Dempster, E., Viana, J. et al. (2016). An
integrated genetic-epigenetic analysis of
schizophrenia: evidence for co-localization of genetic
associations and differential DNA
methylation. Genome Biol, 17, 176.
Kaminsky, Z. A., Tang, T., Wang, S. C., Ptak, C., Oh,
G.H., Wong, A. H., Feldcamp, L. A., Virtanen, C.,
Halfvarson, J., Tysk, C. et al. (2009). DNA
methylation profiles in monozygotic and dizygotic
twins. Nat. Genet., 41, 240–245.
Merikangas, A.K., Corvin, A. P. and Gallagher, L. (2009).
Copy-number variants in neurodevelopmental
disorders: promises and challenges. Trends Genet.,
pp.25, 536–544.
Miklowitz, D. J., & Johnson, S. L. Bipolar disorder. In W.
E. Craighead, D. J. Miklowitz, & L. W. Craighead
(Eds.). (2008). Psychopathology: History, diagnosis,
and empirical foundations, pp.366–401.
Mill, J., Tang, T., Kaminsky, Z., Khare, T., Yazdanpanah,
S., Bouchard, L., Jia, P., Assadzadeh, A., Flanagan, J.,
Schumacher, A. et al. (2008). Epigenomic profiling
reveals DNA-methylation changes associated with
major psychosis. Am. J. Hum. Genet., pp.82, 696–
711.
Patel, V. and Prince, M. (2010). Global mental health: a
new global health field comes of age. JAMA, 303,
1976–1977.
Roy Lardenoije, Artemis Iatrou, Gunter Kenis,
Konstantinos Kompotis, Harry W.M. Steinbusch,
Diego Mastroeni, Paul Coleman, Cynthia A. Lemere,
Patrick R. Hof, Daniel L.A. van den Hove, Bart P.F.
Rutten. (2015). The epigenetics of aging and
neurodegeneration,Progress in Neurobiology, Volume
131, Pages 21-64, ISSN 0301-0082.
Weber, M., Hellmann, I., Stadler, M.B., Ramos, L., Paabo,
S., Rebhan, M. and Schubeler, D. (2007). Distribution,
silencing potential and evolutionary impact of
promoter DNA methylation in the human genome.
Nat. Genet., 39, 457–466.
Influence of Epigenetic Differences on the Etiology of Bipolar Disorder and Schizophrenia
477
