
Correlation between Differential Expression of m6A and Prognosis of
Uterine Corpus Endometrial Carcinoma
Mengqi Zuo
Shanghai Tianjiabing Secondary School, Shanghai, China
Keywords: Uterine Corpus Endometrial Carcinoma (UCEC), Overall Survival (OS), m6A, Prognosis Prediction.
Abstract: UCEC, known as Uterine corpus endometrial carcinoma, is one of the most common types of gynecologic
malignancy worldwide. Notwithstanding great focus has been put on the treatments of UCEC recently, both
the incidence rate and mortality rate of UCEC are still increasing. The 5-year overall survival (OS) rate in
early-stage UCEC ranges from 74 to 91%. Chemotherapy and hormone therapy are viable treatment options
for patients with metastasis or recurrence. However, not all patients benefit from these. For advanced stage
III or IV disease, the 5-year OS rates are 57–66% and 20–26%, respectively. The most common form of post
transcriptional RNA modification, N6-methyladenine (m6A) has attracted increasing interest in cancer
pathogenesis and progression. The differential expression of m6A could be an important clue in the area of
prognosis. Thus, we aimed to identify the correlations between m6A expressions and prognosis of UCEC,
and build a prognostic gene signature in UCEC. In this study, firstly, we filtrated and analysed the gene
expression in RNA sequence and the matched clinical information of UCEC patients from The Cancer
Genome Atlas (TCGA) database. Second, we determined that several m6A regulatory genes had a significant
negative impact on patient survival. By using the Statistical Product and Service Solutions (SPSS) and R-
studio, we built both a univariate Cox regression model and a multivariate Cox regression model. In the end,
we discovered these four m6A gene expressions that had a significant association with the UCEC patient
survival data: VIRMA, METTL14, HNRNPC and FTO. Whereas the multivariate Cox regression model’s
analysis suggested that risk score might be an independent prognostic indicator for the overall survival of
patients with UCEC (p-value ¡0.05). In conclusion, m6A regulator could be an effective and reliable
biomarker for future UCEC prognosis prediction and it deserves further research.
1 INTRODUCTION
Uterine Corpus Endometrial Carcinoma (UCEC), a
common gynaecologic malignancy worldwide, is
defined as an epithelial neoplasm originating from the
endometrium. According to recent research, it is
estimated that there will be 66,570 new cases and an
estimated 12,940 people will die of this disease in
2021 worldwide. Recently, increasing attention has
been paid to adjuvant therapy and targeted therapy in
the overview of the main research progress on UCEC.
Indeed, great advances were made in the treatments
of UCEC. However, the incidence and mortality rates
are still increasing globally. Under this circumstance,
it is crucial to identify novel clinical potential
prognostic biomarkers and therapeutic targets to
improve the patients’ survival of UCEC.
To date, various post-transcriptional RNA
modifications have been discovered and identified as
an epigenetic regulation mechanism in cells and play
a crucial role in a variety of biological diseases,
especially cancers. N6-methyladenine (m6A) mRNA
modification, being the most abundant form of RNA
modification in eukaryotes, has attracted increasing
interest recently. M6A modification relies on a series
of enzymes, which are named “writers”
(methyltransferases), “erasers” (demethylases), and
“readers” (m6A-binding proteins), that can add,
remove, or preferentially bind to m6A functional
sites, thereby altering important biological functions.
The mechanism of m6A in cancer pathogenesis and
progression has been reported in various studies. For
example, researchers found that METTL3, a type of
methyltransferase, acts 2 as an oncogene in lung
cancer and nasopharyngeal carcinoma (NPC).
METTL3 enhances translation of epidermal growth
factor receptor (EGFR). In lung squamous cell
carcinoma, METTL3 interacts with eukaryotic
Zuo, M.
Correlation between Differential Expression of m6A and Prognosis of Uterine Corpus Endometrial Carcinoma.
DOI: 10.5220/0011375000003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 979-984
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
979

translation initiation to accelerate tumorigenicity by
promoting translation of oncogenic mRNAs, such as
Bromodomain-containing protein 4 (BRD4).
In this study, we first analyzed the gene
expression data in RNA sequences and matched
clinical information of UCEC patients from The
Cancer Genome Atlas (TCGA) database. The mRNA
expression levels of a total of 16 m6A regulators were
significantly correlated with different patient data.
Multivariate Cox regression and survival test analysis
suggested that the risk score based on the p-value
(¡0.05) might be an independent prognostic indicator
for the overall survival of patients of UCEC.
2 MATERIALS AND METHODS
2.1 Data Acquisition and Processing
Clinical data of UCEC, including gene expression
RNAseq FPKM, phenotype, and surviving data, were
downloaded from the UCSC Xena website
(https://xenabrowser.net/datapages/). Then they were
processed and sorted by Excel. The download time
was July 2021. There were originally 185 cases
included within the HTseq FPKM data. By using the
VLOOKUP function, the corresponding gene stable
ID was matched between the surviving data and gene
expression data. After excluding the false data and
null data, there were exclusive cases that were closely
correlated and matched with the patient and surviving
data. Each of the 20 m6A gene expression data was
extracted; then they were processed and categorized
into two conditions-low or high gene expression.
Meanwhile, age, overall survival (OS) time, Clinical
M and Clinical T, these 4 categories and their
following data were selected from the phenotype data
acting as another set of variables. Considering periods
usually ended as the patient’s age reached above 45,
the patient age data were differentiated into two
categories: lower or equal to the age of 45, or higher
than the age of 45.
2.2 Proportional Hazard Regression
Model
We denote that 𝑓(𝑡) being the probability density
function (PDF), ℎ(𝑡) being the hazard ratio, and 𝑆(𝑡)
being the survival.
𝑆
(
𝑡
)
=1−𝐹(𝑡) (1)
ℎ
(
𝑡
)
=
()
()
(2)
ℎ
(
𝑡
)
=ℎ
(𝑡) ∙ 𝑒𝑥𝑝
𝑋𝛽
(3)
In this model we do not assume that the hazard
ratio changes by time with each patient. The
assumption is that the hazard ratio is proportional to
each risk group.
After we processed and categorized the data by
using Excel, we used the Statistical Product and
Service Solutions (SPSS) and language 𝑅 to
complete further research. By using the Cox
regression model and the survival analysis, we
discovered associations between data sets. After the
data is analyzed, we can use a function to describe the
risk factor.
𝑌 = 0.781 ×
(
𝑅𝐵𝑀15𝐵
)
+ 0.781 ×
(
𝑉𝐼𝑅𝑀𝐴
)
+
0.878 ×
(
𝐼𝐺𝐹2𝐵𝑃2
)
+ 0.798 ×
(
𝐻𝑁𝑅𝑁𝑃𝐴2𝐵1
)
+
0.622 ×
(
𝐼𝐺𝐹2𝐵𝑃1
)
+ 0.799 ×
(
𝑌𝑇𝐻𝐷𝐹3
)
+ 0.606 ×
(
𝐼𝐺𝐹2𝐵𝑃3
)
+ 0.663 ×
(
𝐻𝑁𝑅𝑁𝑃𝐶
)
+ 0.750 ×
(
𝑅𝐵𝑀15
)
+ 0.686 ×
(
𝑅𝐵𝑀𝑋
)
+ 1.060 ×
(
𝑀𝐸𝑇𝑇𝐿14
)
+
0.793 ×
(
𝑌𝑇𝐻𝐷𝐶2
)
+ 0.721 ×
(
𝑀𝐸𝑇𝑇𝐿3
)
+ 0.947 ×
(
𝑍𝐶2𝐻13
)
+ 0.621 ×
(
𝑊𝑇𝐴𝑃
)
+ 0.584 ×
(
𝑌𝑇𝐻𝐷𝐹1
)
+
1.204 ×
(
𝑌𝑇𝐻𝐷𝐶1
)
+ 0.819 ×
(
𝐹𝑇𝑂
)
+ 0.726 ×
(
𝑌𝑇𝐻𝐷𝐹2
)
+ 0.749 × (𝐴𝐿𝐾𝐵𝐻5) (4)
In this equation, the expression of all twenty m 6A
genes and their hazard ratio are shown.
Then we can determine whether each m6A has a
high or low gene expression. By categorizing each
high or low expression of m6A, we can make a risk
stratification system. In addition, we have added the
clinical data as another variable to judge whether the
gene expression is an independent risk factor itself or
not. We added the patient age, clinical T and clinical
M as three new covariations to the Cox regression
model. After the results came out, we determined that
VIRMA, HNRNPC, METTL14 and FTO were
independent risk factors themselves and were not
influenced by patient data (patient age, clinical T and
clinical M).
3 MATERIALS AND METHODS
Table 1 shows the basic tendency of my data. Those
data based on 1 and 0 were analyzed. Since the OS
time and clinical T are not pure 0s and 1s data, I will
use mean standard deviation to express OS time
clinical T.
Based on the data and the progress in R-studio, I
made this Kaplan-Meier plot. Age=0 means that the
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
980

recorded age is equal to or below 45. Age=1 means
that the recorded age is above 45.
Through the Kaplan-Meier plot, we can clearly
see that patients with age equal to or below 45 have a
higher survival probability. Especially when the time
reached 1000-2500 days of getting UCEC, the two
sets of data drew a significant distance.
Table 1: Variable stratified survival.
0 1 p
n 51 28
RBM15B = 1 (%) 21 (41.2) 18 (64.3) 0.084
VIRMA = 1 (%) 22 (43.1) 17 (60.7) 0.208
IGF2BP2 = 1 (%) 22 (43.1) 17 (60.7) 0.208
HNRNPA2B1 = 1 (%) 19 (37.3) 20 (71.4) 0.008
IGF2BP1 = 1 (%) 19 (37.3) 20 (71.4) 0.008
YTHDF3 = 1 (%) 26 (51.0) 13 (46.4) 0.879
IGF2BP3 = 1 (%) 22 (43.1) 17 (60.7) 0.208
HNRNPC = 1 (%) 19 (37.3) 20 (71.4) 0.008
RBM15 = 1 (%) 16 (31.4) 23 (82.1) <0.001
RBMX = 1 (%) 22 (43.1) 17 (60.7) 0.208
METTL14 = 1 (%) 30 (58.8) 9 (32.1) 0.042
YTHDC2 = 1 (%) 26 (51.0) 13 (46.4) 0.879
METTL3 = 1 (%) 22 (43.1) 17 (60.7) 0.208
ZC2H13 = 1 (%) 27 (52.9) 12 (42.9) 0.534
WTAP = 1 (%) 23 (45.1) 16 (57.1) 0.430
YTHDF1 = 1 (%) 25 (49.0) 14 (50.0) 1.000
YTHDC1 = 1 (%) 26 (51.0) 13 (46.4) 0.879
FTO = 1 (%) 29 (56.9) 10 (35.7) 0.118
YTHDF2 = 1 (%) 22 (43.1) 17 (60.7) 0.208
ALKBH5 = 1 (%) 24 (47.1) 15 (53.6) 0.750
OS.time (mean (SD)) 1765.06 (1087.45) 914.68 (669.02) <0.001
age = 1 (%) 28 (54.9) 16 (57.1) 1.000
ClinicalM = 1 (%) 3 ( 5.9) 12 (42.9) <0.001
ClinicalT (mean (SD)) 2.04 (0.80) 3.04 (1.14) <0.001
Correlation between Differential Expression of m6A and Prognosis of Uterine Corpus Endometrial Carcinoma
981
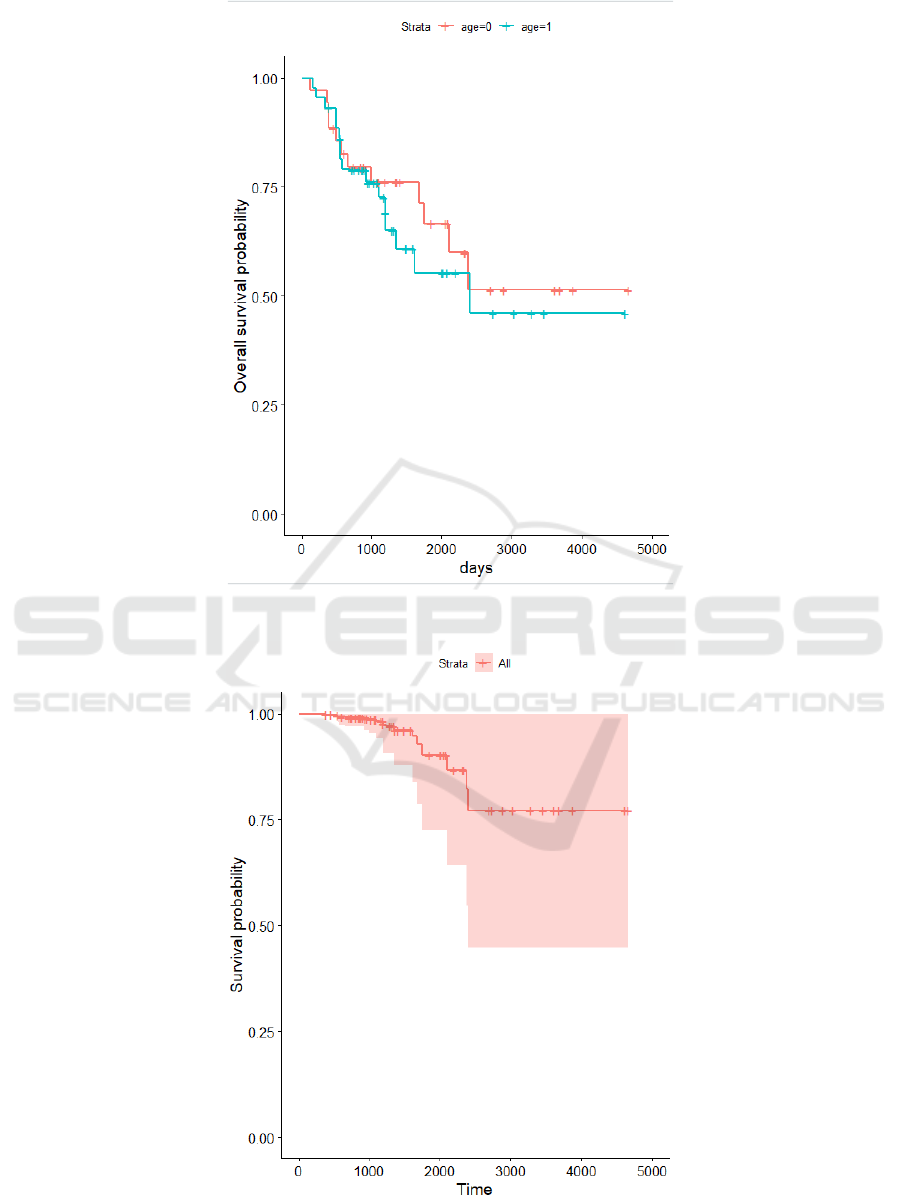
Figure 1: Kaplan Meier Plot.
Figure 2: Proportional hazard model.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
982

Table 2: Proportional hazard model.
coef exp(coef) se(coef)
z Pr(>|z|)
RBM15B 0.33 1.39 0.78
0.42 0.68
VIRMA 1.78 5.92 0.78
2.28 0.02
IGF2BP2 -0.32 0.73 0.88
-0.36 0.36
HNRNPA2B1 1.48 4.41 0.80
1.86 0.06
IGF2BP1 0.93 2.53 0.62
1.49 0.14
YTHDF3 -0.86 0.42 0.80
-1.08 0.28
IGF2BP3 0.33 1.39 0.61
0.54 0.59
HNRNPC 1.78 5.96 0.66
2.69 0.01
RBM15 0.66 1.93 0.75
0.88 0.38
RBMX -1.13 0.32 0.69
-1.64 0.10
METTL14 -2.79 0.06 1.06
-2.63 0.01
YTHDC2 1.15 3.15 0.79
1.45 0.15
METTL3 0.37 1.44 0.72
0.51 0.61
ZC2H13 -0.11 0.89 0.95
-0.12 0.91
WTAP 0.28 1.33 0.62
0.45 0.65
YTHDF1 0.4. 1.49 0.58
0.68 0.50
YTHDC1 1.52 4.55 1.20
1.26 0.21
FTO -2.47 0.08 0.82
-3.02 0.00
YTHDF2 -1.16 0.31 0.73
-1.60 0.11
ALKBH5 0.31 1.36 0.75
0.41 0.68
age 0.04 1.04 0.67
0.06 0.96
ClinicalM -0.18 0.84 0.82
-0.21 0.83
ClinicalT 0.91 2.52 0.39
2.40 0.02
In Table 2, compared with patients who did not
express VRIMA, the hazard ratio of patients who
expressed VRIMA was increased by 5.92.
The hazard ratio increases by 5.92 for the patients
with HNRNPC expression compared to the patients
without HNRNPC expression.
The hazard ratio increases by 5.92 for the patients
with METTL14 expression compared to the patients
without METTL14 expression.
The hazard ratio increases by 5.92 for the patients
with FTO expression compared to the patients
without FTO expression.
4 DISCUSSION AND
CONCLUSIONS
UCEC - Uterine Corpus Endometrial Carcinoma OS
- Overall Survival, defined as the time from
randomization to death from any cause, is a direct
measure of clinical benefit to a patient. It is also a
good standard primary end point to evaluate the
outcome of procedure that is assessed in oncologic
clinical trials.
In conclusion, m6A regulator could be an
effective and reliable biomarker for future UCEC
prognosis prediction and it deserves further research.
Indeed, among the 21 m6A genes, only 4 of them
were closely related to the stage and the risk level.
Correlation between Differential Expression of m6A and Prognosis of Uterine Corpus Endometrial Carcinoma
983

This means the prognosis with m6A still has some
limitations. Also, there are still some uncertainties in
this research. For example, RBMX, IGF2BP1, and
other m6A genes. But that is not to say that it is not
beneficial.
In fact, the number of genes and samples included
in this study are limited. In further study, data
enrichment should be used to have a more accurate
and reliable result. The 4 m6A genes we had
identified their different gene expression having an
impact on patient survival data: VIRMA, METTL14,
HNRNPC and FTO. These 4 m6A genes could be
used as a biomark for the prognosis of UCEC
patients.
REFERENCES
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J,
Du P, Kim W, Tang S, Sliz P, et al. mRNA
circularization by METTL3-eIF3h enhances translation
and promotes oncogenesis. Nature. 201c8; 561: 556–
60.
James J Driscoll, Oliver Rixe, 2009. Overall survival: still
the gold standard: why overall survival remains the
definitive end point in cancer clinical trials. [online]
pubmed. Available at:
https://pubmed.ncbi.nlm.nih.gov/19826360/ [Accessed
29 November 2021].
Murali R., Soslow R.A. and Weigelt B. (2014)
Classification of endometrial carcinoma: more than two
types. Lancet Oncol. 15, e268–e278 10.1016/S1470-
2045(13)70591-6
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, Wu Q, Yuan
B, Lu Q, Yang H. Mechanism of RNA modification
N6-methyladenosine in human cancer. Mol Cancer.
2020 Jun 8;19(1):104. doi: 10.1186/s12943-020-
01216-3. PMID: 32513173; PMCID: PMC7278081.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
984
