
Transcriptional Changes of Genes Linked to Alzheimer’s Disease
Wanning He
a
Shenzhen College of International Education, Futian District, Shenzhen, China
Keywords: Alzheimer’s Disease, GWAS, Bulk RNA-Seq.
Abstract: Alzheimer’s disease is a pogressive neurodegenerative disease that constitutes most cases of dementia. This
study aims to converge existing data from GWAS studies and bulk RNA-Seq from patients with and without
Alzheimer’s disease to prioritize genes involved in the disease pathology. For this study, I examine existing
bulk RNA-Seq datasets from patients with and without Alzheimer’s disease [GSE159699], focusing on genes
previously identified as links to late-onset Alzheimer’s disease in GWAS studies. I confirmed my results with
a publicly available AD transcriptomics consensus tool published by the Swarup lab. In my analysis, I
identified shared gene expression differences in STAG3L5P, MEF2C, MS4A6A, PILRA and CASS4 between
Alzheimer’s disease and control patients across several datasets. These genes were previously linked to late-
onset Alzheimer’s disease. Further investigation should explore how their mutations and gene expression
differences contribute to the mechanisms underlying Alzheimer’s disease.
1 INTRODUCTION
Alzheimer’s disease (AD) was first diagnosed by
Alois Alzheimers in 1907 in a case of a 51-year-old
woman who was experiencing a relatively rapidly
deteriorating memory along with psychiatric
disturbances. Over time, the definition of AD has
changed, and today we consider it to be a neurological
disorder accompanied by a hallmark pathology:
presence of extracellular amyloid beta plaques and
intracellular neurofibrillary tangles formed of tau
protein in the brain (Matthews, Xu, Gaglioti, Holt,
Croft, Mack, McGuire. 2019); (Morgan, 2011).
Currently, AD affects more than 20 million people
worldwide, with about 135 million people expected
to develop it by 2050 (Castellani, Rolston, Smith.
2010); (Fratiglioni, Ronchi, Agüero-Torres. 1999).
AD can be classified into two categories, early
and late-onset, defined by the age of diagnosis and
inheritance pattern (Masters, Bateman, Blennow,
Rowe, Sperling, Cummings. 2015). Whilst early-
onset AD forms around 10% of the cases, around 90%
of AD cases are late-onset, with 85% of the patients
older than 75 years of age (Rabinovici. 2019).
Multiple genetic mutations are responsible for the
development of AD. Mutations in genes processing
amyloid beta proteins are linked to early-onset AD:
a
https://orcid.org/0000-0002-7955-7340
APP, PSEN1 and PSEN2 (Masters, Bateman,
Blennow, Rowe, Sperling, Cummings. 2015).
Genome-wide association studies (GWAS) have
identified a number of risk factors related to late-
onset AD, including the APOE allele e4 (Rabinovici.
2019). APOE e4 carriers have a higher risk of
developing late-onset AD, in contrast to carriers of e2
or e3 alleles. Subsequent GWAS studies have
identified dozens other loci conferring risk factors for
late-onset AD, including TREM2, ADAM10,
ADAMTS1 and others (Kunkle, 2019); (Jansen,
2019).
To understand more about the mechanism behind
the pathology of AD, I evaluated the expressions of
genes linked to late-onset AD in patients with and
without Alzheimer’s disease. I assessed whether the
genes that confer a known risk towards late-onset
Alzheimer’s disease are also differentially expressed
in patients with Alzheimer’s disease, regardless of
their mutation status.
2 METHODOLOGY
Publicly available bulk RNA-Seq datasets from post-
mortem temporal lobes from patients with and
without Alzheimer’s disease were used to investigate
1112
He, W.
Transcriptional Changes of Genes Linked to Alzheimer’s Disease.
DOI: 10.5220/0011378300003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 1112-1118
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
the expression of genes previously identified as
linked to late-onset AD [GSE159699, 9]. The data
was re-analyzed using DESeq2 in Rstudio (Michael,
2014). Six patients that were identified as outliers via
PCA were removed from the dataset, including three
patients with AD and three healthy controls. Using
boxplots, I investigated the expression of genes
previously identified as linked to late-onset AD
(Kunkle, 2019); (Jansen, 2019) in the GSE159699
dataset. To compare my results with previously
published literature, I investigated the expression of
the genes linked to late-onset AD via a publicly
available AD consensus transcriptomics resource
developed by the Swarup lab (Morabito, 2020) which
reanalyzed human AD gene-expression datasets from
several resources, including the ROSMAP dataset
(Bennett, 2018) and the Mayo dataset (Allen, 2016).
The main goal of this study was to investigate the
expression of genes previously identified as
associated with late-onset AD in GWAS studies, in
bulk RNA-Seq data from post-mortem brains with
and without Alzheimer’s disease. For this, I identified
genes of interest associated with late-onset AD
published in recent GWAS studies (Kunkle, 2019);
(Jansen, 2019). Using bulk RNA-Seq dataset from the
temporal lobes of these patients, I compared the
expression of the genes linked to late-onset AD in the
temporal lobes of patients with and without AD
[GSE159699, 9]. To relate my research to previously
published literature, I compared my results to the AD
consensus transcriptomics resources by the Swarup
lab (Nativio, 2020) which was used to display AD
consensus gene expression from multiple sources
(Bennett, 2018); (Allen, 2016).
The GSE159699 dataset is a bulk RNA-Seq
dataset from the temporal lobes of 12 patients with
AD, 10 healthy age-matched controls to the AD
patients and 8 healthy young controls (Nativio, 2020).
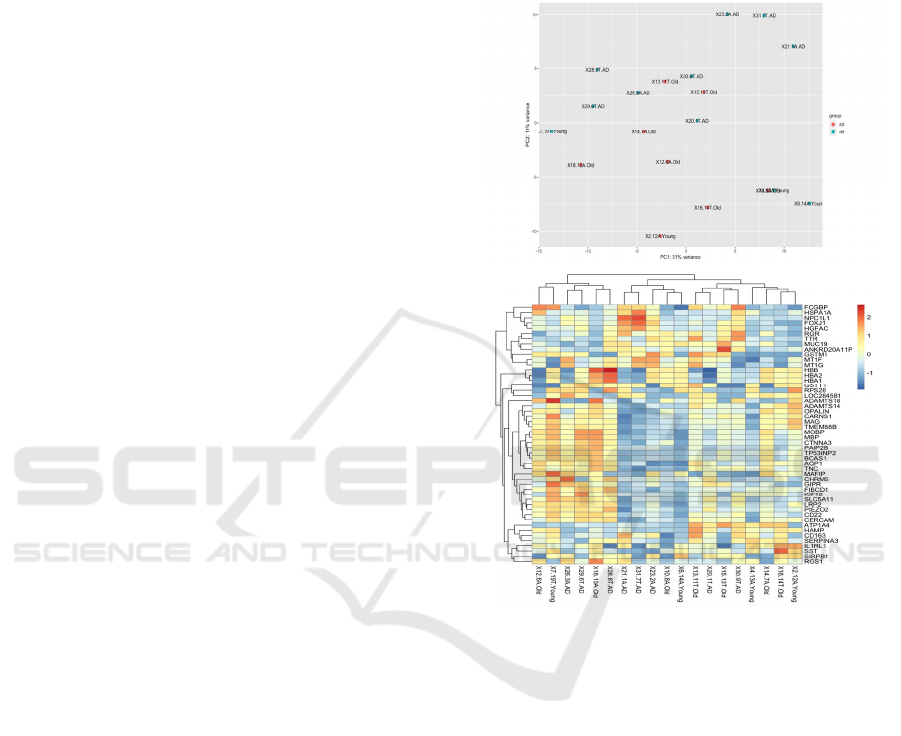
First, I plotted all patients using a PCA plot and a
heatmap of the top 20 differentially expressed genes
in DESeq2 (Michael, 2014). I identified 6 outliers,
that were significantly different from all the other
patients. After removing the outliers from the dataset,
the AD patients (n=9) were separated from the
healthy young (n=4) and healthy old controls (n=7) in
the PCA plot, with PC1 explaining 19% variance in
the dataset (Figure 1). A heatmap of the top 20
differentially expressed genes did not identify any
significant difference between AD patients and the
healthy and young controls. This result was
unremarkable and in line with previous results
published by Nativio et al. (2020), who reported
differences in expression among the genes related to
epigenetic alterations (Nativio, 2020). The original
paper by Nativio et al. (2020) analyzed gene
expression of only genes related to the GO term
‘regulation of transcription’ (Nativio, 2020), whereas
my heatmap graph presented the top 20 genes that
were differentially expressed among the AD, young
and old healthy controls (Figure 1).
Figure 1: PCA and heatmap of top 20 differentially
expressed genes among the bulk RNA-Seq data from the
temporal lobes of patients with and without AD
[GSE159699, 9].
Because genes that are differently expressed
between AD and controls were previously discussed
and reported in the original paper by Nativio et al.
(Nativio, 2020), I did not seek to replicate the analysis,
but instead decided to look into whether genes
previously identified as links to late-onset AD in
GWAS studies are differently expressed between AD
and controls in the GSE159699 bulk RNA-Seq
dataset which contained AD patients, along with old
and healthy controls. For this, I identified genes
linked to late-onset AD using previously published
GWAS studies (Kunkle, 2019); (Jansen, 2019). For
each gene previously identified as linked to late-onset
AD, I reported whether it was differently expressed in
the current dataset. In addition, I crossed my results
with the AD transcriptomics consensus resource
Transcriptional Changes of Genes Linked to Alzheimer’s Disease
1113
developed by the Swarup lab (Morabito, 2020). This
resource allows plotting of gene expression
differences among the brains of AD patients, age-
matched patients, and healthy controls. The plots
below summarize fold-change expressions between
AD and controls using the GSE159699 dataset
(Nativio, 2020), and the AD consensus resource by
the Swarup lab (Morabito, 2020).
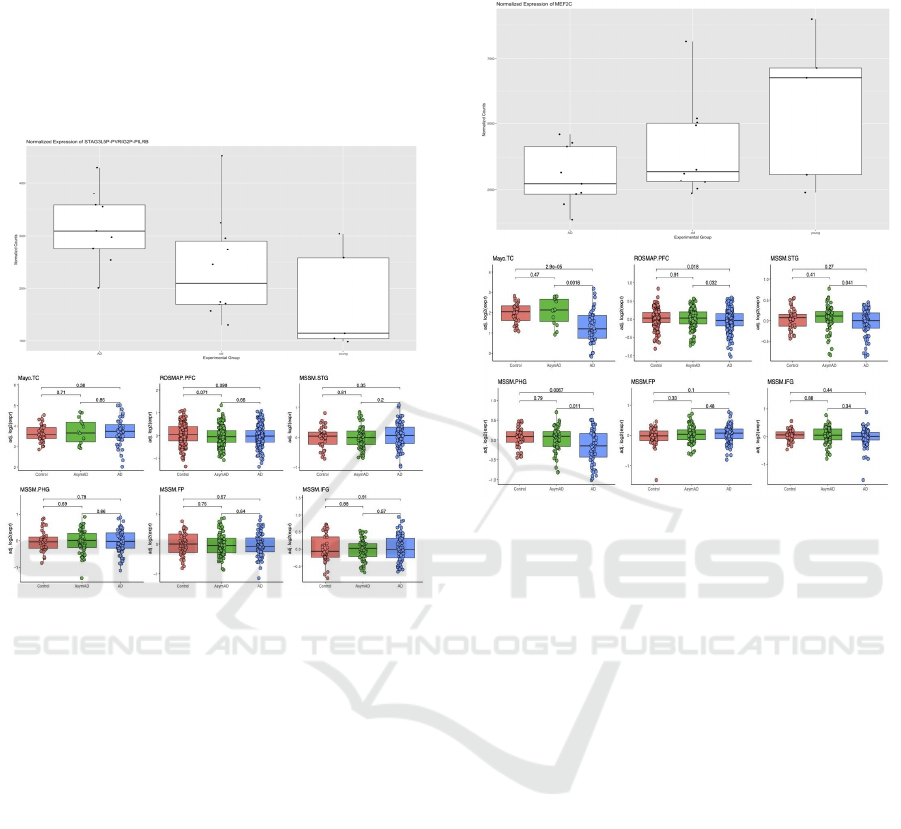
Figure 2: The boxplot of differences in STAG3L5P-
PVRIG2P-PIL gene expression among AD, along with
healthy young and old controls indicates a significant
increase of expression of STAG3L5P - PVRIG2P - PIL in
the AD group. On the left is the boxplot of gene expression
differences plotted using the bulk RNA-Seq data from the
temporal lobes of patients with and without AD
[GSE159699, 9]. On the right is the boxplot of gene
expression differences in bulk RNA-Seq datasets using the
AD gene expression consensus resource developed by the
Swarup lab (Morabito, 2020). Three published datasets
show an upregulated expression while three shows a
downregulated expression.
The STAG3L5P gene displays an increase in
expression in AD vs control patients in the
GSE159699 dataset. When compared to the AD
consensus transcriptomics resource by the Swarup lab,
STAG3L5P also showed an increase in expression in
the ROSMAP dataset (Fig.7a). Previously, the
exome-sequencing studies showed that the
associations with two variants in a novel gene STAG3
were also replicated and significantly associated with
AD in the replication analysis. The rare variants in
STAG3 identified by WGS suggested the possibility
that STAG3 has a distinct mechanistic role in AD
which is different from other normal variants (Joshua,
2020).
Figure 3: The boxplot of differences in ME2FC gene
expression among AD patients, along with healthy young
and old controls indicates a significant decrease in ME2FC
of expression in the AD group. On the left is the boxplot of
gene expression differences plotted using the bulk RNA-
Seq data from the temporal lobes of patients with and
without AD [GSE159699, 9]. On the right is the boxplot of
gene expression differences in bulk RNA-Seq datasets
using the AD gene expression consensus resource
developed by the Swarup lab (Morabito, 2020).
A Similar process was performed for the MEF2C
gene, which showed a decrease in expression in AD
vs control patients in the GSE159699 dataset.
Comparison to the AD consensus resource by the
Swarup lab revealed unclear changes in gene
expression between AD patients and the controls,
displaying a decreased expression in AD brains using
the Mayo and the MSMM dataset, but not in the other
datasets (Figure 3). MEF2C has a role in conferring
resilience to pro-inflammatory stimuli in microglia
(Deczkowska, 2017). Microglia plays an important
role in AD, and MEF2C restricts the microbial
response to immune stimuli. Additionally, other
GWAS studies show that mutations in MEF2C are
linked to late-onset AD. Inflammation is known to be
associated withcognitive dysfunction and may
contribute to the pro-inflammatory milieu of the brain
in AD or aging patients (Simen, 2011).
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1114

MS4A6A displays an increased expression in the
AD group using the GSE159699 dataset, Mayo and
MSSM dataset, but the gene expression relationship
is unclear using the other datasets from the AD
consensus resource by the Swarup lab (Nativio, 2020)
(Morabito, 2020). Previous studies show that
MS4A6A is associated with AD and is likely to have
an immune-related function (Reitz. 2015); (Paul,
2011).
Figure 4: The boxplot of differences in MS4A6A gene
expression among AD patients along with healthy young
and old controls indicates an increase in MS4A6A of
expression in the AD group. On the left is the boxplot of
gene expression differences plotted using the bulk RNA-
Seq data from the temporal lobes of patients with and
without AD [GSE159699, 9]. On the right is the boxplot of
gene expression differences in bulk RNA-Seq datasets
using the AD gene expression consensus resource
developed by the Swarup lab (Morabito, 2020).
The PILRA gene is associated with late-onset AD
and exhibits an increase in gene expression in the AD
vs control group in the GSE159699 dataset and the
MSSM datasets from the AD consensus resource by
the Swarup lab. Previous research revealed a
significant burden of PILRA variants in the exome-
wide burden analysis of AD (Patel, 2018).
Figure 5: The boxplot of differences in PILRA gene
expression among AD patients along with healthy young
and old controls indicates an increase in PILRA expression
in the AD group. On the left is the boxplot of gene
expression differences plotted using the bulk RNA-Seq data
from the temporal lobes of patients with and without AD
[GSE159699, 9]. On the right is the boxplot of gene
expression differences in bulk RNA-Seq datasets using the
AD gene expression consensus resource developed by the
Swarup lab (Morabito, 2020).
Figure 6: The boxplot of differences in CASS4 gene
expression among AD patients along with healthy young
and old controls indicates an increase in CASS4 expression
in the AD group. On the left is the boxplot of gene
expression differences plotted using the bulk RNA-Seq data
from the temporal lobes of patients with and without AD
[GSE159699, 9]. On the right is the boxplot of gene
expression differences in bulk RNA-Seq datasets using the
AD gene expression consensus resource developed by the
Swarup lab (Morabito, 2020).
The CASS4 gene exhibits an increase in
expression in AD patients vs the control group in the
GSE159699 dataset as well as the Mayo dataset using
the AD consensus resource from the Swarup lab.
CASS4 was previously found to be associated with
the amyloid, tau pathology, cytoskeletal function and
Transcriptional Changes of Genes Linked to Alzheimer’s Disease
1115

the axonal transport pathways identified in GWAS
studies (Reitz. 2015). It was found to retain the motifs
required for the interaction with PTK2B and to
contribute to the pathology of AD (Beck, 2014).
3 DISCUSSIONS
AD is a progressive neurodegenerative disease that
accounts for the most cases of dementia. This study
aimed to converge existing data from GWAS studies
and RNA-Seq to prioritize the high-risk genes for AD.
I found that 5 genes, STAG3L5P, MEF2C, MS4A6A,
PILRA and CASS4, were both linked to AD in the
GWAS studies and differentially expressed in the
RNA-Seq analysis between the AD and control
groups using the GSE159699 dataset (Nativio, 2020).
To validate my findings, I compared my results with
an AD transcriptomics consensus tool published by
the Swarup lab (Morabito, 2020), that reanalyzed
golden-standard bulk RNA-Seq datasets from AD
patients along with healthy old and young controls
using various datasets, including Mayo and
ROSMAP (Bennett, 2018); (Allen, 2016).
4 CONCLUSIONS
My analysis shows that STAG3L5P, MEF2C,
MS4A6A, PILRA and CASS4 exhibit changes in
expression between AD and control patients that are
fairly consistent across different datasets. These
results suggest that these genes could be particularly
important in AD. All these genes were previously
found to be associated with AD in GWAS studies
(Kunkle, 2019); (Jansen, 2019). In addition, their
gene functions are relevant to AD mechanisms.
STAG3LAP has several rare variants identified
through exome-wide analysis, with suspected distinct
mechanisms in causing AD (Joshua, 2020). MS4A6A
has a gene function associated with microglial
function and immunity (Reitz. 2015); (Hollingworth,
2011). The five genes explored in this study should
be further investigated to confirm their causality to
AD, and the role of the genetic variants and changes
in the expression of mechanisms leading to AD.
Further understanding of how STAG3L5P,
MEF2C, MS4A6A, PILRA and CASS4 contribute to
AD may be used for early detection, prevention and
drug development in AD.
ACKNOWLEDGEMENTS
If any, should be placed before the references section
without numbering.
REFERENCES
Aleksandra Deczkowska, Orit Matcovitch-Natan, Afroditi
Tsitsou-Kampeli, Sefi Ben-Hamo, Raz Dvir-
Szternfeld, Amit Spinrad, Oded Singer, Eyal David,
Deborah R. Winter, Lucas K. Smith, Alexander
Kertser, Kuti Baruch, Neta Rosenzweig, Anna Terem,
Marco Prinz, Saul Villeda, Ami Citri, Ido Amit, and
Michal Schwartz. 2017. Mef2C restrains microglial
inflammatory response and is lost in brain ageing in an
IFN-I-dependent manner. Nature Communications 8, 1:
717. https://doi.org/10.1038/s41467-017-00769-0
Arthur A. Simen, Kelly A. Bordner, Mark P. Martin,
Lawrence A. Moy, and Lisa C. Barry. 2011. Cognitive
Dysfunction with Aging and the Role of Inflammation.
Therapeutic Advances in Chronic Disease 2, 3: 175–
195. https://doi.org/10.1177/2040622311399145
Colin L. Masters, Randall Bateman, Kaj Blennow,
Christopher C. Rowe, Reisa A. Sperling, and Jeffrey L.
Cummings. 2015. Alzheimer’s disease. Nature
Reviews Disease Primers 1, 1: 1–18.
https://doi.org/10.1038/nrdp.2015.56
Christiane Reitz. 2015. Genetic diagnosis and prognosis of
Alzheimer’s disease: challenges and opportunities.
Expert review of molecular diagnostics 15, 3: 339–348.
https://doi.org/10.1586/14737159.2015.1002469
D. Morgan. 2011. Immunotherapy for Alzheimer’s disease.
Journal of Internal Medicine 269, 1: 54–63.
https://doi.org/10.1111/j.1365-2796.2010.02315.x
David A. Bennett, Aron S. Buchman, Patricia A. Boyle,
Lisa L. Barnes, Robert S. Wilson, and Julie A
Schneider. 2018. Religious Orders Study and Rush
Memory and Aging Project. Journal of Alzheimer’s
disease: JAD 64, Suppl 1: S161–S189.
https://doi.org/10.3233/JAD-179939
Gil D. Rabinovici. 2019. Late-onset Alzheimer Disease.
Continuum: Lifelong Learning in Neurology 25, 1: 14–
33. https://doi.org/10.1212/CON.0000000000000700
Iris E. Jansen, Jeanne E. Savage, Kyoko Watanabe, Julien
Bryois, Dylan M. Williams, Stacy Steinberg, Julia
Sealock, Ida K. Karlsson, Sara Hägg, Lavinia
Athanasiu, Nicola Voyle, Petroula Proitsi, Aree
Witoelar, Sven Stringer, Dag Aarsland, Ina S. Almdahl,
Fred Andersen, Sverre Bergh, Francesco Bettella,
Sigurbjorn Bjornsson, Anne Brækhus, Geir Bråthen,
Christiaan de Leeuw, Rahul S. Desikan, Srdjan
Djurovic, Logan Dumitrescu, Tormod Fladby, Timothy
J. Hohman, Palmi V. Jonsson, Steven J. Kiddle, Arvid
Rongve, Ingvild Saltvedt, Sigrid B. Sando, Geir
Selbæk, Maryam Shoai, Nathan G. Skene, Jon Snaedal,
Eystein Stordal, Ingun D. Ulstein, Yunpeng Wang,
Linda R. White, John Hardy, Jens Hjerling-Leffler,
Patrick F. Sullivan, Wiesje M. van der Flier, Richard
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1116

Dobson, Lea K. Davis, Hreinn Stefansson, Kari
Stefansson, Nancy L. Pedersen, Stephan Ripke, Ole A.
Andreassen, and Danielle Posthuma. 2019. Genome-
wide meta-analysis identifies new loci and functional
pathways influencing Alzheimer’s disease risk. Nature
Genetics 51, 3: 404–413.
https://doi.org/10.1038/s41588-018-0311-9
Joshua C. Bis, Xueqiu Jian, Brian W. Kunkle, Yuning
Chen, Kara L. Hamilton-Nelson, William S. Bush,
William J. Salerno, Daniel Lancour, Yiyi Ma, Alan E.
Renton, Edoardo Marcora, John J. Farrell, Yi Zhao,
Liming Qu, Shahzad Ahmad, Najaf Amin, Philippe
Amouyel, Gary W. Beecham, Jennifer E. Below,
Dominique Campion, Laura Cantwell, Camille
Charbonnier, Jaeyoon Chung, Paul K. Crane, Carlos
Cruchaga, L. Adrienne Cupples, Jean-François
Dartigues, Stéphanie Debette, Jean-François Deleuze,
Lucinda Fulton, Stacey B. Gabriel, Emmanuelle Genin,
Richard A. Gibbs, Alison Goate, Benjamin Grenier-
Boley, Namrata Gupta, Jonathan L. Haines, Aki S.
Havulinna, Seppo Helisalmi, Mikko Hiltunen, Daniel
P. Howrigan, M. Arfan Ikram, Jaakko Kaprio, Jan
Konrad, Amanda Kuzma, Eric S. Lander, Mark
Lathrop, Terho Lehtimäki, Honghuang Lin, Kari
Mattila, Richard Mayeux, Donna M. Muzny, Waleed
Nasser, Benjamin Neale, Kwangsik Nho, Gaël Nicolas,
Devanshi Patel, Margaret A. Pericak-Vance, Markus
Perola, Bruce M. Psaty, Olivier Quenez, Farid Rajabli,
Richard Redon, Christiane Reitz, Anne M. Remes,
Veikko Salomaa, Chloe Sarnowski, Helena Schmidt,
Michael Schmidt, Reinhold Schmidt, Hilkka Soininen,
Timothy A. Thornton, Giuseppe Tosto, Christophe
Tzourio, Sven J. van der Lee, Cornelia M. van Duijn,
Otto Valladares, Badri Vardarajan, Li-San Wang,
Weixin Wang, Ellen Wijsman, Richard K. Wilson,
Daniela Witten, Kim C. Worley, Xiaoling Zhang,
Celine Bellenguez, Jean-Charles Lambert, Mitja I.
Kurki, Aarno Palotie, Mark Daly, Eric Boerwinkle,
Kathryn L. Lunetta, Anita L. Destefano, Josée Dupuis,
Eden R. Martin, Gerard D. Schellenberg, Sudha
Seshadri, Adam C. Naj, Myriam Fornage, and Lindsay
A. Farrer. 2020. Whole exome sequencing study
identifies novel rare and common Alzheimer’s-
Associated variants involved in immune response and
transcriptional regulation. Molecular Psychiatry 25, 8:
1859–1875. https://doi.org/10.1038/s41380-018-0112-
7
Kevin A. Matthews, Wei Xu, Anne H. Gaglioti, James B.
Holt, Janet B. Croft, Dominic Mack, and Lisa C.
McGuire. 2019. Racial and ethnic estimates of
Alzheimer’s disease and related dementias in the
United States (2015–2060) in adults aged ≥65 years.
Alzheimer’s & dementia: the journal of the Alzheimer’s
Association, 15(1), 17–24.
https://doi.org/10.1016/j.jalz.2018.06.3063
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C.,
Damotte, V., Naj, A. C., Boland, A., Vronskaya, M.,
van der Lee, S. J., Amlie-Wolf, A., Bellenguez, C.,
Frizatti, A., Chouraki, V., Martin, E. R., Sleegers, K.,
Badarinarayan, N., Jakobsdottir, J., Hamilton-Nelson,
K. L., Moreno-Grau, S., . . . Pericak-Vance, M. A.
(2019b). Genetic meta-analysis of diagnosed
Alzheimer’s disease identifies new risk loci and
implicates Aβ, tau, immunity and lipid processing.
Nature Genetics, 51(3), 414–430.
https://doi.org/10.1038/s41588-019-0358-2
L. Fratiglioni, D. De Ronchi, and H. Agüero-Torres. 1999.
Worldwide prevalence and incidence of dementia.
Drugs & Aging 15, 5: 365–375.
https://doi.org/10.2165/00002512-199915050-00004
Michael I. Love, Wolfgang Huber, and Simon Anders.
2014. Moderated estimation of fold change and
dispersion for RNA-seq data with DESeq2. Genome
Biology 15, 12: 550. https://doi.org/10.1186/s13059-
014-0550-8
Mariet Allen, Minerva M. Carrasquillo, Cory Funk,
Benjamin D. Heavner, Fanggeng Zou, Curtis S.
Younkin, Jeremy D. Burgess, High-Seng Chai, Julia
Crook, James A. Eddy, Hongdong Li, Ben Logsdon,
Mette A. Peters, Kristen K. Dang, Xue Wang, Daniel
Serie, Chen Wang, Thuy Nguyen, Sarah Lincoln,
Kimberly Malphrus, Gina Bisceglio, Ma Li, Todd E.
Golde, Lara M. Mangravite, Yan Asmann, Nathan D.
Price, Ronald C. Petersen, Neill R. Graff-Radford,
Dennis W. Dickson, Steven G. Younkin, and Nilüfer
Ertekin-Taner. 2016. Human whole genome genotype
and transcriptome data for Alzheimer’s and other
neurodegenerative diseases. Scientific Data 3: 160089.
https://doi.org/10.1038/sdata.2016.89
Paul Hollingworth, Denise Harold, Rebecca Sims, Amy
Gerrish, Jean-Charles Lambert, Minerva M.
Carrasquillo, Richard Abraham, Marian L. Hamshere,
Jaspreet Singh Pahwa, Valentina Moskvina, Kimberley
Dowzell, Nicola Jones, Alexandra Stretton, Charlene
Thomas, Alex Richards, Dobril Ivanov, Caroline
Widdowson, Jade Chapman, Simon Lovestone, John
Powell, Petroula Proitsi, Michelle K. Lupton, Carol
Brayne, David C. Rubinsztein, Michael Gill, Brian
Lawlor, Aoibhinn Lynch, Kristelle S. Brown, Peter A.
Passmore, David Craig, Bernadette McGuinness,
Stephen Todd, Clive Holmes, David Mann, A. David
Smith, Helen Beaumont, Donald Warden, Gordon
Wilcock, Seth Love, Patrick G. Kehoe, Nigel M.
Hooper, Emma R. L. C. Vardy, John Hardy, Simon
Mead, Nick C. Fox, Martin Rossor, John Collinge,
Wolfgang Maier, Frank Jessen, Eckart Rüther, Britta
Schürmann, Reiner Heun, Heike Kölsch, Hendrik van
den Bussche, Isabella Heuser, Johannes Kornhuber,
Jens Wiltfang, Martin Dichgans, Lutz Frölich, Harald
Hampel, John Gallacher, Michael Hüll, Dan Rujescu,
Ina Giegling, Alison M. Goate, John S. K. Kauwe,
Carlos Cruchaga, Petra Nowotny, John C. Morris,
Kevin Mayo, Kristel Sleegers, Karolien Bettens,
Sebastiaan Engelborghs, Peter P. De Deyn, Christine
Van Broeckhoven, Gill Livingston, Nicholas J. Bass,
Hugh Gurling, Andrew McQuillin, Rhian Gwilliam,
Panagiotis Deloukas, Ammar Al-Chalabi, Christopher
E. Shaw, Magda Tsolaki, Andrew B. Singleton, Rita
Guerreiro, Thomas W. Mühleisen, Markus M. Nöthen,
Susanne Moebus, Karl-Heinz Jöckel, Norman Klopp,
Transcriptional Changes of Genes Linked to Alzheimer’s Disease
1117

H.-Erich Wichmann, V. Shane Pankratz, Sigrid B.
Sando, Jan O. Aasly, Maria Barcikowska, Zbigniew K.
Wszolek, Dennis W. Dickson, Neill R. Graff-Radford,
Ronald C. Petersen, Alzheimer’s Disease
Neuroimaging Initiative, Cornelia M. van Duijn,
Monique M. B. Breteler, M. Arfan Ikram, Anita L.
DeStefano, Annette L. Fitzpatrick, Oscar Lopez,
Lenore J. Launer, Sudha Seshadri, CHARGE
consortium, Claudine Berr, Dominique Campion,
Jacques Epelbaum, Jean-François Dartigues,
Christophe Tzourio, Annick Alpérovitch, Mark
Lathrop, EADI1 consortium, Thomas M. Feulner,
Patricia Friedrich, Caterina Riehle, Michael Krawczak,
Stefan Schreiber, Manuel Mayhaus, S. Nicolhaus,
Stefan Wagenpfeil, Stacy Steinberg, Hreinn
Stefansson, Kari Stefansson, Jon Snaedal, Sigurbjörn
Björnsson, Palmi V. Jonsson, Vincent Chouraki,
Benjamin Genier-Boley, Mikko Hiltunen, Hilkka
Soininen, Onofre Combarros, Diana Zelenika, Marc
Delepine, Maria J. Bullido, Florence Pasquier, Ignacio
Mateo, Ana Frank-Garcia, Elisa Porcellini, Olivier
Hanon, Eliecer Coto, Victoria Alvarez, Paolo Bosco,
Gabriele Siciliano, Michelangelo Mancuso, Francesco
Panza, Vincenzo Solfrizzi, Benedetta Nacmias, Sandro
Sorbi, Paola Bossù, Paola Piccardi, Beatrice Arosio,
Giorgio Annoni, Davide Seripa, Alberto Pilotto, Elio
Scarpini, Daniela Galimberti, Alexis Brice, Didier
Hannequin, Federico Licastro, Lesley Jones, Peter A.
Holmans, Thorlakur Jonsson, Matthias
Riemenschneider, Kevin Morgan, Steven G. Younkin,
Michael J. Owen, Michael O’Donovan, Philippe
Amouyel, and Julie Williams. 2011. Common variants
at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and
CD2AP are associated with Alzheimer’s disease.
Nature Genetics 43, 5: 429–435.
https://doi.org/10.1038/ng.803
Rudy J. Castellani, Raj K. Rolston, and Mark A. Smith.
2010. Alzheimer Disease. Disease-a-month: DM 56, 9:
484–546.
https://doi.org/10.1016/j.disamonth.2010.06.001
Raffaella Nativio, Yemin Lan, Greg Donahue, Simone
Sidoli, Amit Berson, Ananth R. Srinivasan, Oksana
Shcherbakova, Alexandre Amlie-Wolf, Ji Nie,
Xiaolong Cui, Chuan He, Li-San Wang, Benjamin A.
Garcia, John Q. Trojanowski, Nancy M. Bonini, and
Shelley L. Berger. 2020. An integrated multi-omics
approach identifies epigenetic alterations associated
with Alzheimer’s disease. Nature Genetics 52, 10:
1024–1035. https://doi.org/10.1038/s41588-020-0696-
0
Samuel Morabito, Emily Miyoshi, Neethu Michael, and
Vivek Swarup. 2020. Integrative genomics approach
identifies conserved transcriptomic networks in
Alzheimer’s disease. Human Molecular Genetics 29,
17: 2899–2919. https://doi.org/10.1093/hmg/ddaa182
T. Patel, K. J. Brookes, J. Turton, S. Chaudhury, T. Guetta-
Baranes, R. Guerreiro, J. Bras, D. Hernandez, A.
Singleton, P. T. Francis, J. Hardy, and K. Morgan.
2018. Whole-exome sequencing of the BDR cohort:
evidence to support the role of the PILRA gene in
Alzheimer’s disease. Neuropathology and Applied
Neurobiology 44, 5: 506–521.
https://doi.org/10.1111/nan.12452
Tim N. Beck, Emmanuelle Nicolas, Meghan C. Kopp, and
Erica A. Golemis. 2014. Adaptors for disorders of the
brain? The cancer signaling proteins NEDD9, CASS4,
and PTK2B in Alzheimer’s disease. Oncoscience 1, 7:
486–503.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1118
