
Changes of Eukaryotic Microorganism Structures in Soil during
Lettuce-spinach Rotation
Qingwen Li
1a
, Xinyu Wang
1b
, Jie Hong
1c
, Yi Gao
1d
, Zhidi Chen
1e
, Lianquan Zhong
2f
,
Xinxin Yi
1,* g
and Xiuzhi Gao
1,* h
1
Beijing Laboratory of Food Quality and Safety, Beijing Key Laboratory of Agricultural Product Detection and Control of
Spoilage Organisms and Pesticide Residue, Beijing Engineering Laboratory of Probiotics Key Technology Development,
College of Food Science and Engineering, Beijing University of Agriculture, Beijing, 102206, China
2
Beijing Changping District Plant Protection and Plant Quarantine Station, Beijing, 102200, China
zhonglianquan1@163.com,
*
gxz@bua.edu.cn,
*
yixinxin2008@163.com
*
Corresponding author
Keywords:
Eukaryotic Microorganism, Lettuce-spinach Rotation, High-Throughput Sequencing Technology.
Abstract:
The purpose of this study is to analyze and compare eukaryotic microorganisms in soil samples by rotation of
lettuce and spinach. High-throughput sequencing technology was used to analyze the eukaryotes present in
soil samples before and after crop planting and the effect of soil sucrase activity on eukaryotic communities,
in order to provide data that will aid in alleviating the problems resulting from the continuous cropping of
lettuce. The results showed that eukaryotic microorganism, such as Heterodera, Pseudallescheria and
Podosphaera species, were inhibited by rotation of lettuce with spinach and had a certain effect on the
recovery of soil quality. Rotation of lettuce with spinach could increase lettuce production and soil sucrase
activity by 31.4% and 9.5%, respectively. It could improve the diversity of eukaryotic microbial community
in soil.
1 INTRODUCTION
Soil microbial diversity is important for sustainable
agricultural development, because microorganisms
can participate in several biochemical processes that
promote agricultural production, including plant
nutrient recycling, soil structure maintenance and
agricultural chemical degradation (Lanzén 2013). Soil
microorganisms are mainly composed of bacteria,
fungi, actinomycetes and some algae. Among them,
soil eukaryotes play an important role in maintaining
soil nutrients and biogeochemical cycles. There are
many factors affecting soil microbial community,
including soil characteristics, environmental
a
https://orcid.org/0000-0003-1882-8351
b
https://orcid.org/0000-0002-4304-4738
c
https://orcid.org/0000-0002-9672-9554
d
https://orcid.org/0000-0002-6981-4032
e
https://orcid.org/0000-0003-3894-0252
f
https://orcid.org/0000-0001-8295-4294
g
https://orcid.org/0000-0002-2139-1149
h
https://orcid.org/0000-0002-1122-4742
conditions, crop management strategies and other
factors, among which plant species and tillage are
important factors affecting soil microbial community
(Larkin, 2003). Soil enzyme activity is also an
important indicator of microbial and biochemical
processes, usually involving the decomposition and
synthesis of soil organic matter, nutrient cycling and
availability, soil fertility and quality, and its
determination can be used to quantify and monitor
changes in microbial community structure and
activity, and soil Dynamic response index of organic
matter to human disturbance (Raiesi, 2018).
Researchers such as Lawton and Li found that the
overall composition of species and microbial
1254
Li, Q., Wang, X., Hong, J., Gao, Y., Chen, Z., Zhong, L., Yi, X. and Gao, X.
Changes of Eukaryotic Microorganism Structures in Soil during Lettuce-spinach Rotation.
DOI: 10.5220/0011393000003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 1254-1259
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

communities and planting methods with certain
functional characteristics could change the structure
and diversity of soil microbial communities and
affected ecosystem functions (Lawton, 1994, Li, Liu,
2017).
Lettuce is one of the most popular raw leafy
vegetables in China. So far, there have been few
reports on the interaction of eukaryotic
microorganisms in the soil during lettuce cultivation
and with different planting methods. In soil
microbiology research, soil quality indicators have
been obtained by measuring changes in microbial
activity and community structure over time. In this
paper, the soil collection of the rotation of lettuce and
spinach was used to analyze the changes in soil
eukaryotic microbial community diversity under this
planting method, in order to provide some theoretical
basis for alleviating the obstacle of continuous
cropping of lettuce.
2 MATERIALS AND METHODS
2.1 Site Description
The experiments were conducted in a plastic
greenhouse in an experimental demonstration base
(east longitude 116.14°, north latitude 40.19°) in
Beijing, China. Prior to the experiments, mung beans
were grown within the greenhouse for a long period
of time. The annual average temperature in this area
was 12.6 ℃, and the annual precipitation was 680.6
mm.
2.2 Experimental Design
The experimental setup the lettuce-spinach rotation
group (S). The sample naming format was used S -
planting year – cultivation number - 1 (before
planting) / 2 (after planting) - soil depth. The adjacent
lands were protected by two furrows that were 1.2 m
wide and 6.5 m in length. The experiments were
conducted between September 2016 and June 2017.
Due to low temperatures during the winter, the
experimental field used for the continuous cropping
of lettuce was filled in, and no crops were planted
during this period. Other treatments were consistent
throughout the planting period. Randomly selected
20 × 20 cm area and the yield of lettuce in this area
was weighed by weighing method. Each field soil
sample was collected using a five-point sampling
method, during which four samples were collected at
each of four corners, and one sample was collected
from the center of the field; samples were collected
before and after crop cultivation at sampling depths
of 0-10 cm and 10-20 cm. After the removal of
residual leaves and roots, the soil samples were
placed into aseptic sampling bags. The samples from
each of the five points were combined and divided
into two parts: one part was used for the experiments,
while the other was stored at -40 ℃ for follow-up
experiments. A description of each of the soil samples
is given in Table 1.
Table 1: Description of soil samples.
Sample Collection date Depth (cm) State of crop growth Cultivation time
S.16.1.1.10 2016.09.09 0-10 Before cultivation 1st
S.16.1.1.20 2016.09.09 10-20 Before cultivation 1st
S.16.1.2.10 2016.10.20 0-10 Harvest 1st
S.16.1.2.20 2016.10.20 10-20 Harvest 1st
S.16.2.1.10 2016.10.27 0-10 Before cultivation 2nd
S.16.2.1.20 2016.10.27 10-20 Before cultivation 2nd
S.17.2.2.10 2017.03.10 0-10 Harvest 2nd
S.17.2.2.20 2017.03.10 10-20 Harvest 2nd
S.17.3.1.10 2017.03.21 0-10 Before cultivation 3rd
S.17.3.1.20 2017.03.21 10-20 Before cultivation 3rd
S.17.3.2.10 2017.04.27 0-10 Harvest 3rd
S.17.3.2.20 2017.04.27 10-20 Harvest 3rd
S.17.4.1.10 2017.05.23 0-10 Before cultivation 4th
S.17.4.1.20 2017.05.23 10-20 Before cultivation 4th
S.17.4.2.10 2017.06.20 0-10 Harvest 4th
S.17.4.2.20 2017.06.20 10-20 Harvest 4th
Changes of Eukaryotic Microorganism Structures in Soil during Lettuce-spinach Rotation
1255
2.3
DNA Extraction and PCR
Amplification
According to the manufacturer's instructions, the
Mag-Bind® Universal Metagenomics Kit was used
to extract DNA from 1.0 g of soil samples. The
extracted DNA was measured by agarose gel
electrophoresis (0.8%), and the DNA was quantified
by an ultraviolet spectrophotometer. The extracted
DNA was stored at -80°C for analysis. The V4-V5
region within the 18S rRNA gene was amplified from
each sample using general eukaryotic primers
TAReuk454F WD1 (5'-
CCAGCASCYGCGGTAATTCC-3') and TAReu
kREV3 (5'-ACTTTCGTTCTTGATYRA-3')
(Logares, R 2016) according to previously published
protocols. PCR amplification was conducted using
the Q5 high fidelity DNA polymerase (NEB, UK);
Strictly control the number of amplification cycles to
ensure that the number of cycles was as few as
possible, and the amplification conditions of each
batch of samples were consistent. The high-
throughput sequencing of the 18S rRNA gene was
conducted using the Illumina MiSeq PE300 platform
at the Shanghai Majorbio Bio-pharm Biotechnology
Co., Ltd. (Shanghai, China). The read sequences were
deposited into the NCBI Sequence Read Archive
under accession numbers SRP155301 and
SRP154689.
2.4
Sequence Analysis
In order to integrate the original double-end
sequencing data into our analysis, the sliding window
method was used to individually screen double-end
sequences in FASTQ format. The FLASH software
(v1.2.7; http://ccb.jhu.edu/ software/FLASH/) was
used to pair double-ended sequences through primary
quality screening of overlapping bases. The
sequencing results were analyzed using the QIME
software (v1.8.0; http://qiime.org/). Sequences that
met the following criteria were filtered out: (1)
length<150 bp; (2) contained fuzzy bases; (3) number
of mismatched bases in 5'-end primers > 1; (4)
number of consecutive identical bases > 8. Chimeric
sequences were verified and removed using
USEARCH (v5.2.236;
http://www.drive5.com/usearch/). The QIIME and
UCLUST softwares were used to divide the
operational taxonomic units (OTU) at 97% similarity;
The most abundant sequence in each OTU was
selected as the representative sequence of the OTU.
Then, according to the number of sequences
corresponding to each OTU in each sample, a matrix
file containing the abundance of OTU in each sample
was constructed. For each OTU representative
sequence, used the default parameters in the QIIME
software to compare the representative sequence with
the template sequence in the Silva database to obtain
the classification information corresponding to each
OTU (Release 115; http://www.arb-silva.de).
3 RESUITS
3.1 Difference in Lettuce Yield and Soil
Sucrase Activity
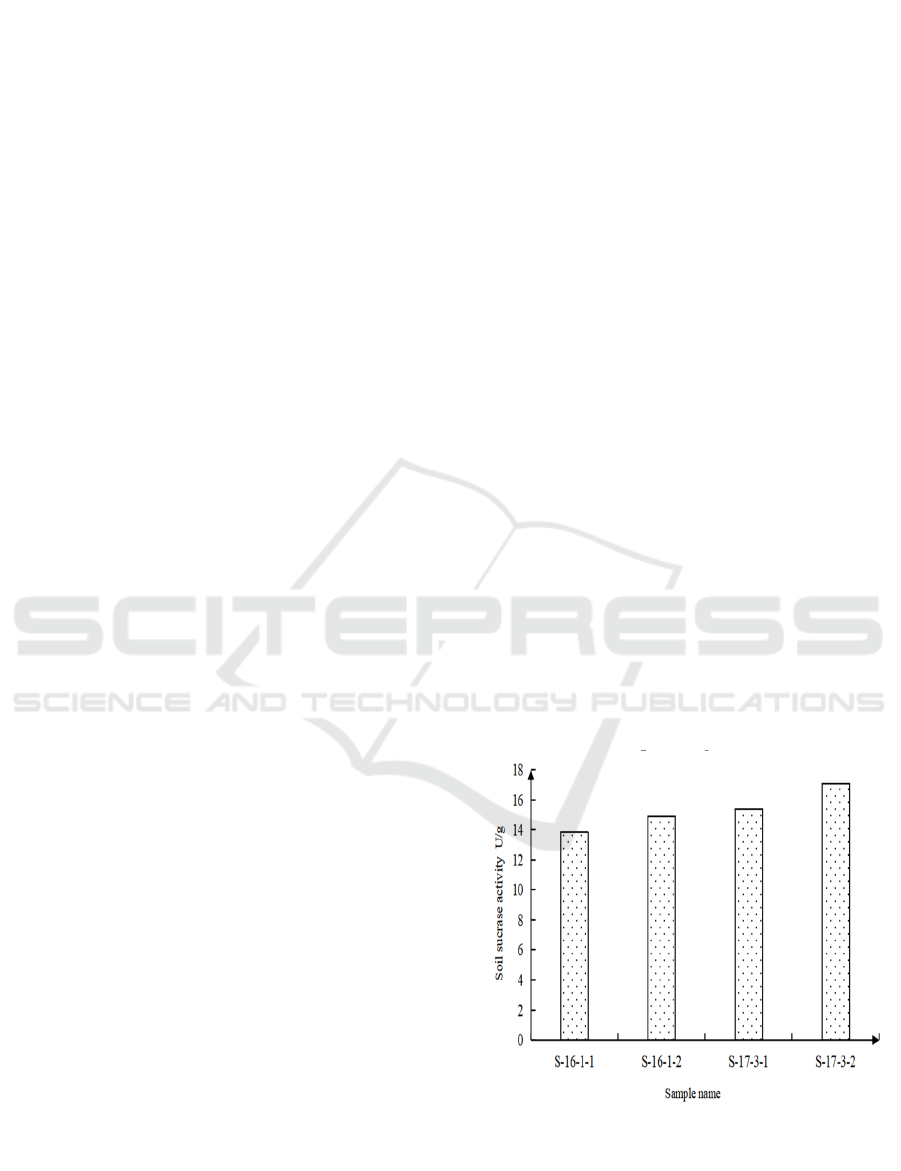
Lettuce grown in rotation with spinach yields were
4.78 kg/m2 and 6.28 kg/m2. The average yield of the
two rotations is 5.53 kg/m2. The output of lettuce
increased by 31.4% after rotation, significantly. In the
process of lettuce rotation, the activity of invertase
fluctuates greatly, and the overall level remained at
15.25 U/g. Sucrose activity increased when lettuce
was harvested compared to pre-planting activity. The
soil invertase activity in rotation treatment increased
by 9.5% on average compared with that before
planting. (Fig. 1). In the crop rotation soil samples,
Ascomycota, Chlorophyta, Nematoda, Streptophyta,
Apicomplexa, Chytridiomycota, Arthropoda and
Eustigmatophyceae accounted for a large proportion
of the eukaryotes phyla present during cultivation. In
addition, Eustigmatophyceae appeared during lettuce
cultivation, and Bacillariophyta was found in all of
the 0-10 cm soil samples (Fig. 2).
Figure 1: Soil sucrase activity in rotation cropping soil
samples.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1256
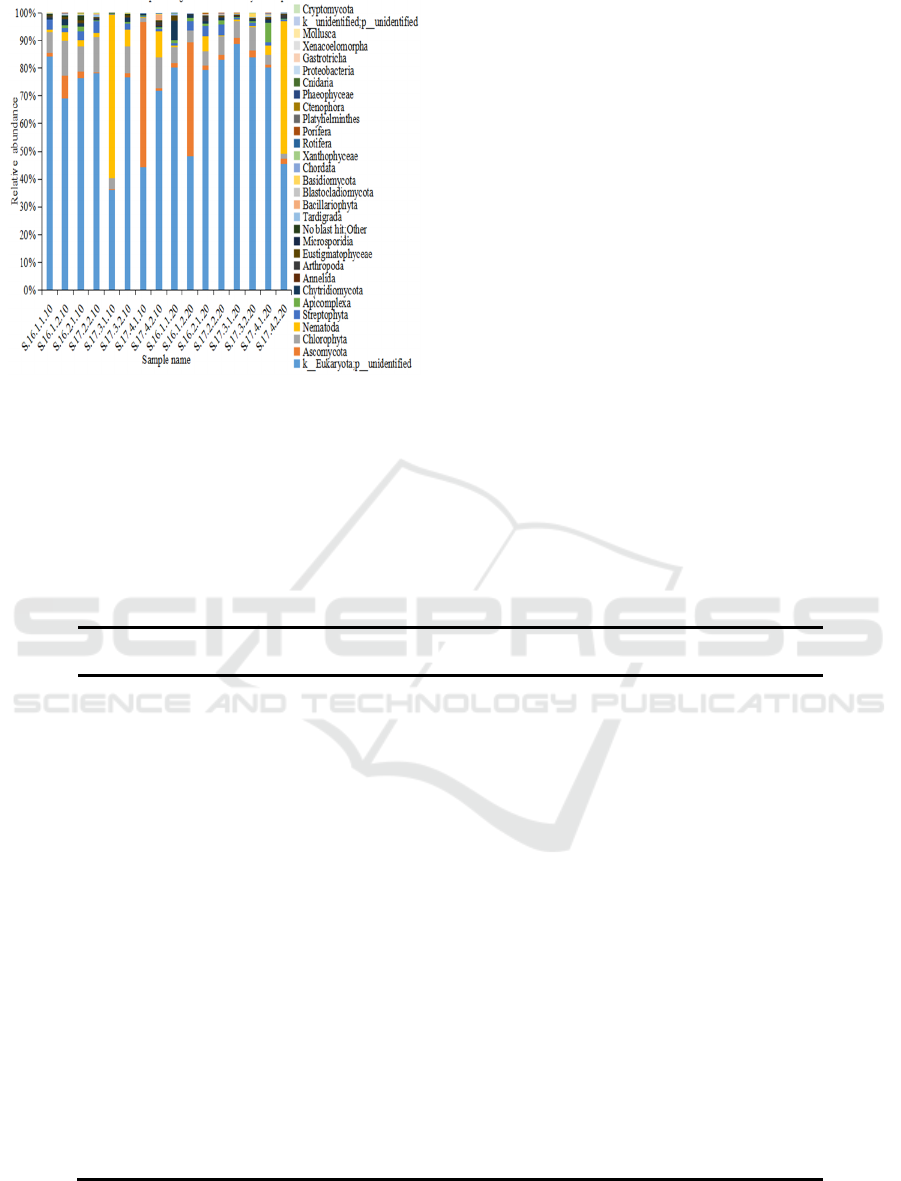
Figure 2: Relative abundance of the eukaryotes phyla
presents in the crop rotation soil samples. Relative
abundances are reported as the percentage of the total
eukaryotes sequences.
3.2 Soil Eukaryotes Diversity and
Community Structure during Crop
Rotation
In the 0-10 cm soil samples, the diversity indices
showed a rising trend during the cultivation of lettuce,
with increases observed in the Chao1
(13.46~65.04%), ACE (14.05~63.79%), Simpson
(2.90~50.35%) and Shannon (18.55~108.24%)
indices. The diversity indices, including Chao1 (-
12.14%), ACE (-12.21%), Simpson (-2.17%), and
Shannon (-13.19%), decreased during the first
cultivation of spinach. During the second spinach
planting, the diversity indices, including Chao1
(36.95%), ACE (36.85%), Simpson (9.86%) and
Shannon (32.35%), were increased. In the 10-20 cm
soil samples, the diversity indices during lettuce
cultivation were increased (Chao1, 6.48~11.89%;
ACE, 6.35~12.97%; Simpson, -3.5~ -3.11%;
Shannon, -10.70~ -9.52%. During the spinach
planting period, all of the indices, including Chao1
(17.55~24.31%), ACE (16.75~22.56%), Simpson (-
8.70~3.88%) and Shannon (-25.12~21.55%), were
increased (Table 2).
Table 2: The proportion of the shared and unique OTUs in the harvest and cultivation soil samples from each cultivation
period.
Samples Chao1 ACE Simpson Shannon
Goods_coverag
e
Evenness
S.16.1.1.10 900.4000 880.5166 0.9421 5.5672 0.9978 0.5788
S.16.1.2.10 1021.5543 1004.2072 0.9694 6.6001 0.9980 0.6694
S.16.2.1.10 1068.1728 1085.3210 0.9722 6.8789 0.9986 0.6860
S.17.2.2.10 938.4615 952.7641 0.9511 5.9713 0.9991 0.6055
S.17.3.1.10 670.1648 681.4558 0.6239 2.8731 0.9981 0.3126
S.17.3.2.10 1106.0067 1116.1486 0.9381 5.9829 0.9977 0.5978
S.17.4.1.10 820.0132 821.0723 0.8845 5.2434 1.0000 0.5417
S.17.4.2.10 1123.0038 1123.6392 0.9718 6.9342 1.0000 0.6843
S.16.1.1.20 998.0000 998.0000 0.9645 6.9054 1.0000 0.6931
S.16.1.2.20 1116.6429 1127.4693 0.9307 6.2483 0.9994 0.6173
S.16.2.1.20 831.0000 843.8775 0.9412 5.9878 0.9983 0.6239
S.17.2.2.20 1033.0339 1034.2674 0.9777 7.2781 0.9999 0.7269
S.17.3.1.20 939.9874 958.8076 0.9654 6.6445 0.9987 0.6752
S.17.3.2.20 1000.9198 1019.7156 0.9354 5.9335 0.9991 0.5961
S.17.4.1.20 878.6081 882.2762 0.9696 7.0500 0.9993 0.7227
S.17.4.2.20 1032.8430 1030.0534 0.8852 5.2790 0.9977 0.5347
Changes of Eukaryotic Microorganism Structures in Soil during Lettuce-spinach Rotation
1257
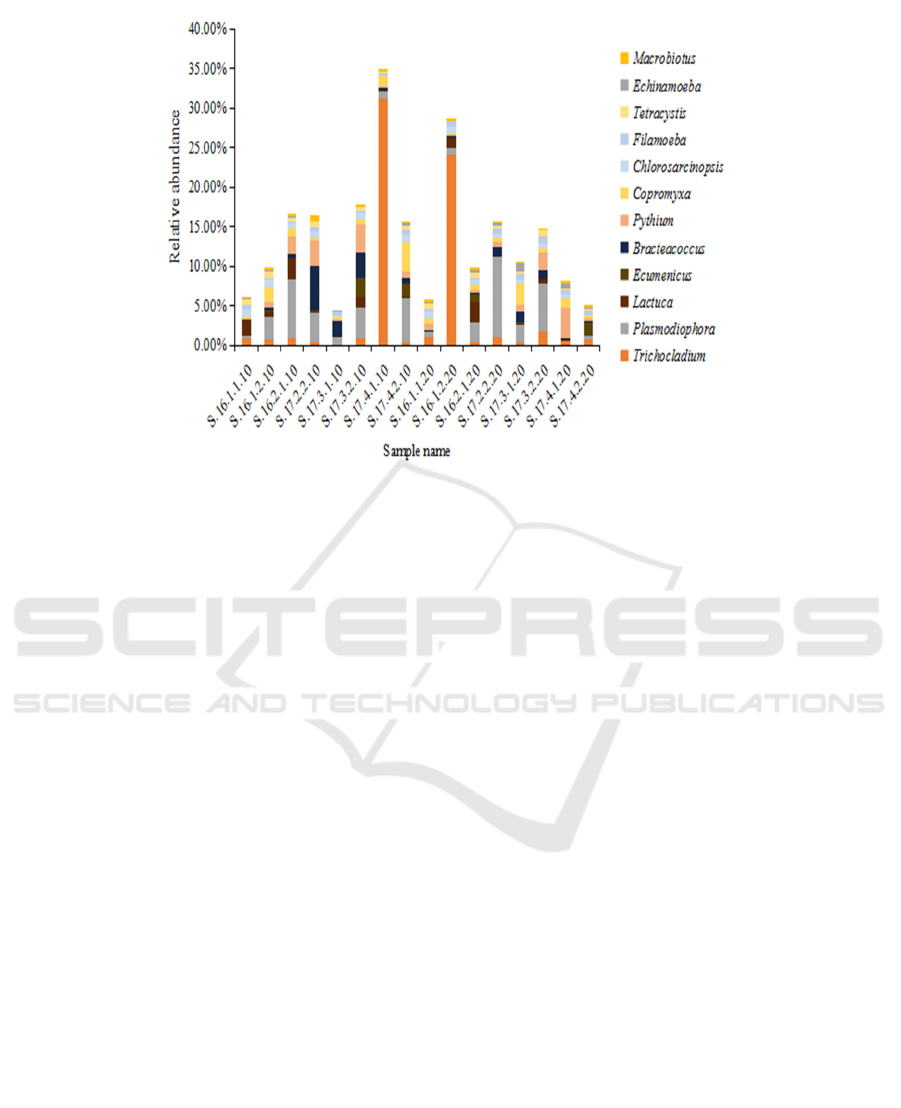
Figure 3: Relative abundance of different eukaryotic genera in the crop rotation soil samples
During crop rotation, Ecumenicus,
Bracteacoccus, Pythium, Chlorosarcinopsis,
Tetracystis and Macrobiotus species showed an
upward trend in the 0-10 cm soil samples obtained
during the cultivation of lettuce and spinach. In the
10-20 cm soil samples, Trichocladium,
Plasmodiophora and Filamoeba species showed an
increasing trend during the rotation process, while
Copromyxa and Echinamoeba species showed a
downward trend. There were also a number of
eukaryotic genera that demonstrated opposing trends
during the cultivation of lettuce and spinach, such as
Lactuca, whose species increased in number during
lettuce cultivation but decreased during the spinach
planting period in the 10-20 cm soil samples (Fig. 3).
4 DISCUSSION
Sucrase is an important enzyme to characterize the
biological activity intensity of soil, which is
positively correlated with the content of humus,
organic matter and clay, the content of N and P, the
number of microorganisms and the respiration
intensity (
Wang, 2005
). In this experiment, the activity
of sucrase and the yield of lettuce both increased
9.5% and 31.4%, respectively, after rotation.
Nematoda were the most dominant faunal group in
this experiment, which is highly diverse, ranked third
in terms of richness during these experiments. In the
soil samples obtained during crop rotation, the
changes in Nematoda were different due to the
different crops that were planted. Among them,
Heterodera species showed an opposing trend during
the process of lettuce and spinach cultivation.
In this experiment, changes in the genus of some
fungi were also different, these differences in
eukaryotic community compositions might be the
result of the different crops. Some studies have found
that members of Ascomycota are the main soil fungal
decomposers (
Ma, 2013
). Interestingly, in the crop
rotation soil samples, this phylum was increased
during lettuce cultivation and decreased during
spinach cultivation, due to different planting
methods. After the lettuce-spinach rotation the
relative abundance of genus such as Bacillus,
Pseudomonas, Sphingomonas, Nitrospira and Zephyr
in soil is higher than the crop rotation. But the the
relative abundance of Acidocaldarius was going low.
Research findings after rotation, the proportion of
bacteria in the soil, such as Acidbacteria, Firmicutes
and Fusarium, is less than that of crop rotation soil.
And Proteobacteria and Bacillus constitute the
dominant flora in the soil in rotation (
Pieterse, 2014
).
Pseudomonas and its secondary metabolites,
including the antibiotic 2,4-diacetylphloroglucinol
(DAPG), protect plants directly by inhibiting plant
pathogens by inducing plant systemic resistance
(
Zhang, 2018
).
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1258

5 CONCLUSIONS
The results showed that crop rotation of spinach and
lettuce could effectively control increased in
Heterodera, Pseudallescheria and Podosphaera
species. However, some eukaryotes species, such as
those of Fusarium and Plasmodiophora, did not
decrease during crop rotation. Further experiments
were needed to identify the specific components of
spinach root exudates that have inhibitory effects on
eukaryotes. In addition, rotation with spinach
improved soil sucrase activity and lettuce yield.
ACKNOWLEDGEMENTS
This work was supported by the Beijing Leafy
Vegetables Innovation Team of Modern Agro-
industry Technology Research System (BAIC07-
2021) and fund for Academic Degree & Graduate
Education of Beijing University of Agriculture
(2021YJS029).
REFERENCES
Lanzén Anders, Addis S, Amare G, et al.(2013) Surprising
prokaryotic and eukaryotic diversity, community
structure and biogeography of ethiopian soda lakes.
Plos One, 8(8): e72577.
Larkin R P. (2003) Characterization of soil microbial
communities under different potato cropping systems
by microbial population dynamics, substrate
utilization, and fatty acid profiles. Soil Biology and
Biochemistry, 35(11): 1451-1466.
Lawton, J H. (1994) What do species do in ecosystems.
Oikos. 71: 367-374. DOI: 10.2307 /3545824.
Li, T, Liu, T and Zheng, C, et al. (2017) Changes in soil
bacterial community structure as a result of
incorporation of brassica plants compared with
continuous planting eggplant and chemical disinfection
in greenhouses. PloS ONE. 12: e0173923. DOI:
10.1371/journal.pone.0173923
Logares, R, Audic, S and Santini, S, et al. (2016) Diversity
patterns and activity of uncultured marine
heterotrophic flagellates unveiled with
pyrosequencing. ISME J.,6(10): 1823-1833. DOI:
10.1038/ismej.2012.36.
Ma, A, Zhuang, X and Wu, J, et al. (2013) Ascomycota
members dominate fungal communities during straw
residue decomposition in arable soil. PLoS ONE. 8:
e66146. DOI: 10.1371/ journal. pone.0066146.
Pieterse, Corne M.J, Zamioudis C, et al. (2014) Induced
systemic resistance by beneficial microbes. Annual
Review of Phytopathology, 52(1): 347-375. DOI:
10.1146/annurev-phyto-082712-102340.
Raiesi F, Salek-Gilani S. (2018) The potential activity of
soil extracellular enzymes as an indicator for ecological
restoration of rangeland soils after agricultural
abandonment. Applied Soil Ecology, 126: 140-147.
Wang H Y. (2005) Study on soil microorganism and Soil
enzyme activity of different vegetation restoration in
the upper reaches of Jialingjiang River. Journal of Soil
and Water Conservation,22(3): 172-177. DOI:
10.3321/j.issn:1009-2242.2008.0 3.035.
Zhang, X, Y, Duan, X, Q and W, M, L et al. (2018) Effects
of rotation and continuous cropping on soil microflora
and diversity in tobacco field. Soil and Fertilizer
Sciences in China. 6: 84-90. DOI:
10.11838/sfsc.20180612.
Changes of Eukaryotic Microorganism Structures in Soil during Lettuce-spinach Rotation
1259
