
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view
Landmarking
Tibor Kub
´
ık
1
and Michal
ˇ
Span
ˇ
el
1,2 a
1
Department of Computer Graphics and Multimedia, Faculty of Information Technology,
Brno University of Technology, Brno, Czech Republic
2
TESCAN 3DIM, Brno, Czech Republic
Keywords:
Landmark Detection in 3D, Polygonal Meshes, Multi-view Deep Neural Networks, RANSAC, U-Net,
Heatmap Regression, Teeth Detection, Dental Scans.
Abstract:
Landmark detection is frequently an intermediate step in medical data analysis. More and more often, these
data are represented in the form of 3D models. An example is a 3D intraoral scan of dentition used in or-
thodontics, where landmarking is notably challenging due to malocclusion, teeth shift, and frequent teeth
missing. What’s more, in terms of 3D data, the DNN processing comes with high memory and computational
time requirements, which do not meet the needs of clinical applications. We present a robust method for
tooth landmark detection based on a multi-view approach, which transforms the task into a 2D domain, where
the suggested network detects landmarks by heatmap regression from several viewpoints. Additionally, we
propose a post-processing based on Multi-view Confidence and Maximum Heatmap Activation Confidence,
which can robustly determine whether a tooth is missing or not. Experiments have shown that the combination
of Attention U-Net, 100 viewpoints, and RANSAC consensus method is able to detect landmarks with an error
of 0.75 ± 0.96 mm. In addition to the promising accuracy, our method is robust to missing teeth, as it can
correctly detect the presence of teeth in 97.68% cases.
1 INTRODUCTION
The localization of landmarks plays a crucial role in
many tasks related to image analysis in medicine.
Deep learning has demonstrated great success in this
field, outperforming conventional machine learning
methods. With the widespread availability of accu-
rate 3D scanning devices, this task has moved into
a 3D domain. This brings us the possibility of in-
creased automation of clinical application tasks that
operate on 3D models, such as in the case of digital
orthodontics.
In terms of direct 3D data processing by neu-
ral networks, a noticeable challenge has emerged as
the size of the input feature vector substantially in-
creases. The time of computation of such deep neu-
ral networks is not suitable for clinical applications
used during treatment planning in digital orthodon-
tics. 3D medical data analysis reckons with an-
other challenge – the limited amount of medical data,
a common struggle in medical image processing.
a
https://orcid.org/0000-0003-0193-684X
Dentition casts used in digital orthodontics soft-
ware are typically obtained from patients with various
levels of malocclusion and numerous kinds of teeth
shifting. Another challenging problem in this do-
main is the absence of teeth, a common phenomenon
in terms of human dentition. The 3rd Molars (also
known as Wisdom teeth) are worth taking a look at.
Their extraction is one of the most frequent proce-
dures in oral surgery as it eliminates future problems
due to unfavorable orientation (Normando, 2015).
Thus, the method should be robust to such variations.
In this paper, we present a method that consid-
ers the limitation of the dataset size, the need for low
computational time, and the importance of robustness
to missing and shifted teeth. It is based on a multi-
view approach and it uses heatmap regression to pre-
dict landmarks in 2D and the RANSAC consensus
method to robustly propagate the information back
into 3D space. In order to address the problem of es-
timation of landmarks on missing teeth, our method
comprises a post-processing based on a heatmap re-
gression uncertainty analysis combined with the un-
certainty of the multi-view approach.
24
Kubík, T. and Špan
ˇ
el, M.
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking.
DOI: 10.5220/0010770700003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 24-34
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
0.0
mm
2.0 mm
4.0 mm
6.0 mm
8.0 mm
10.0+ m
m
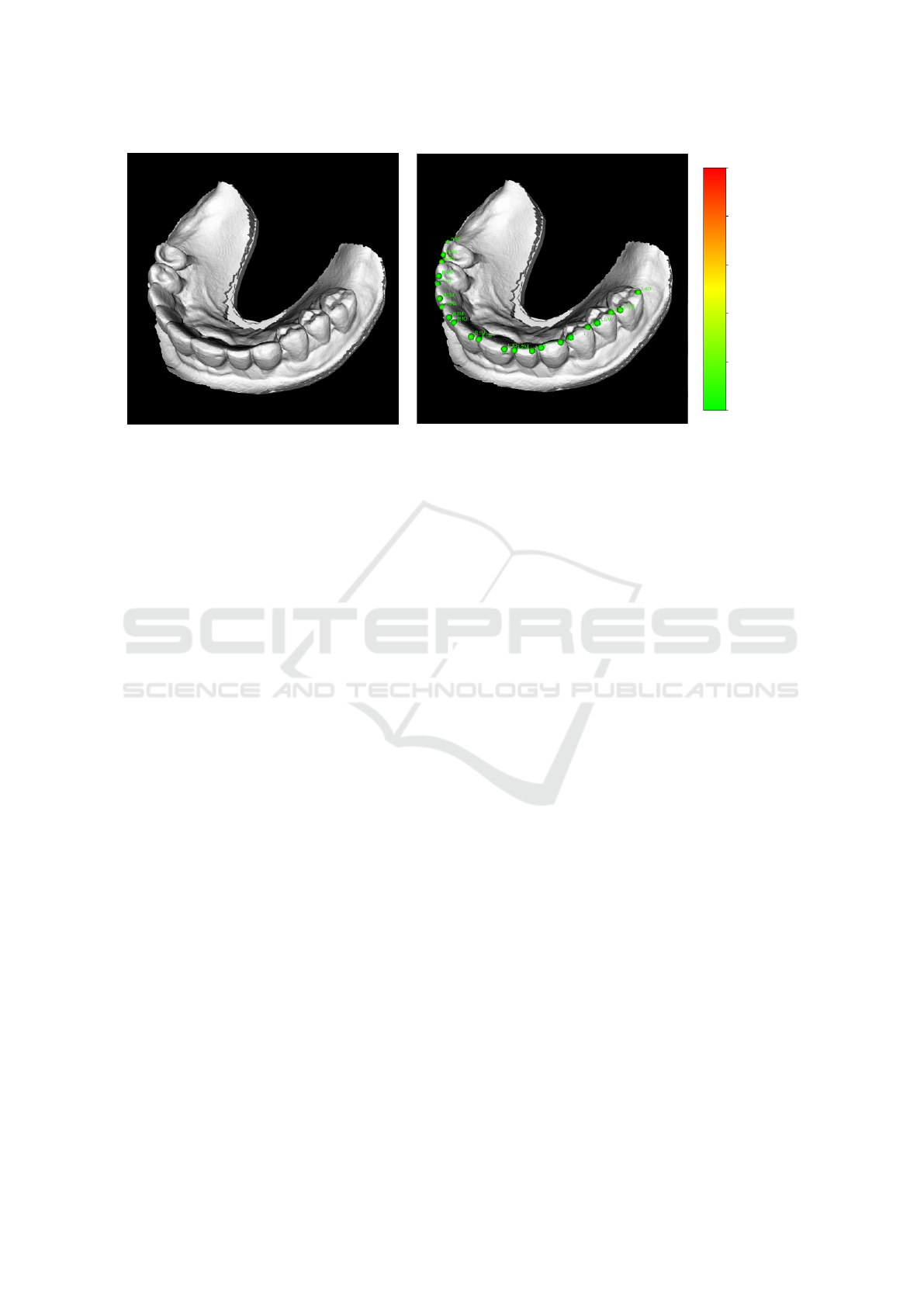
(a) Input polygonal mesh (b) Detected landmarks with our method
Figure 1: An example of a 3D scan of dentition (a) and appropriate detected landmarks (b). Our method automatically detects
two landmarks on each tooth – mesial and distal. This type of landmarks is important in orthodontics, as it defines the rotation
of teeth from anatomically perfect arrangement. Whatsmore, the method correctly detects whether a tooth is missing or not.
Conducted experiments have shown that the pro-
posed method can detect orthodontics landmarks on
surface models with an error of 0.75 ± 0.96 mm
while 98.07% of detected landmarks achieve an er-
ror less than 2 mm. As for the robustness to missing
teeth, our method’s post-processing correctly detects
missing teeth in 97.68% of cases.
2 CURRENT APPROACHES TO
LANDMARKING
Early studies in this area relied on conventional ma-
chine learning approaches. Hough forests were used
for landmark detection. Authors in (Donner et al.,
2013) combined regression and classification, which
brought better results comparing to both a single
voxel’s classification and classification of the vol-
ume of interest. As convolutional neural networks
(CNNs) gained in popularity, an increasing number
of scientific papers concerning their usage in land-
mark detection emerged. Some of these methods de-
tected the landmark position directly by regressing
its x and y coordinates. For example, in (Sun et al.,
2013), the authors adopted cascaded convolutional
neural networks for facial point detection. Another
study (Lv et al., 2017) proposed a regression in a two-
stage manner, still locating landmarks directly.
2.1 Heatmaps in Landmarking
Over time, extensive literature has developed on land-
marking by heatmap regression. The authors in (Pfis-
ter et al., 2015) worked on a model that regresses hu-
man joint positions. Instead of directly regressing the
(x, y) joint position, they regressed a joint position’s
heatmap. During the training, the ground truth labels
are transformed into heatmaps by placing a Gaussian
with fixed variance at each joint coordinate.
On top of the appliance of spatial fusion layers and
optical flow, they discussed the benefits of regress-
ing a heatmap rather than (x, y) coordinates directly.
They concluded that the benefits are twofold: (i) the
process of network training can be visualized in such
a way that one can understand the network learning
failures, and (ii) the network output can acquire confi-
dence at multiple spatial locations. The incorrect ones
are slowly suppressed later in the training process. In
contrast, regressing the (x, y) coordinates directly, the
network would have a lower loss only if it predicts
the coordinate correctly, even if it was “growing con-
fidence” in the correct position. Concerning these,
such an approach outperformed direct coordinate re-
gression and became a standard way of landmark de-
tection in 2D images.
This approach seemed alluring for people in the
medical image processing community. Inspired by
this method, authors in (Payer et al., 2016) pre-
sented multiple architectures that detect keypoints in
X-Ray images of hands and 3D hand MR scans. They
affirmed that by regressing heatmaps, it is possible
to achieve state-of-the-art localization performance in
2D and 3D domains while dealing with medical data
shortage.
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking
25

2.2 Processing of 3D Data by Neural
Networks
Although the extension of deep neural network op-
erations such as convolution from 2D to 3D domain
seems natural, the additional computational complex-
ity introduces notable challenges. Having volumet-
ric data (for example, voxel models) or 3D surface
data (for example, represented as polygon meshes) as
an input to deep neural networks has a considerable
drawback in computational time and memory require-
ments.
An alternative way of 3D data processing by neu-
ral networks is the multi-view approach. Obtaining
state-of-the-art results on 3D classification, authors
in (Su et al., 2015) presented the multi-view CNN
idea. It is relatively straightforward and consists of
three main steps:
1. Render a 3D shape into several images using vary-
ing camera extrinsics.
2. Extract features from each acquired view.
3. Process features from different viewpoints in
a way suitable for a given task. In (Su et al., 2015),
a pooling layer followed by fully connected layers
was used to get class predictions.
The multi-view approach was later on used to
identify feature points on facial surfaces (Paulsen
et al., 2018). The authors discussed multiple geom-
etry derivatives and experimented with their combi-
nations to bring state-of-the-art results in feature point
detection on facial 3D scans while decreasing the pro-
hibitive GPU memory requirements needed for true
3D processing. Additionally, they proposed a con-
sensus method to find the final estimate, which com-
bines the least-squares fit and RANdom SAmple Con-
sensus (RANSAC) (Fischler and Bolles, 1981). For
each landmark, N rays in 3D space are the outputs of
the proposed method.
Based on Graph Neural Networks (GNNs), au-
thors in (Sun et al., 2020) presented coupled 3D seg-
mentation for annotation of individual teeth and gin-
giva. Their network produces a dense correspondence
that helps to accurately locate individual orthodon-
tics landmarks on teeth crowns. Another recent work
in landmark localization on dental mesh models was
presented by authors in (Wu et al., 2021). They in-
troduced a two-stage framework based on mesh deep
learning (TS-MDL) for joint tooth labeling and land-
mark identification. To accurately detect tooth land-
marks, they designed a modified PointNet (Qi et al.,
2017) to learn the heatmaps encoding landmark loca-
tions.
We have developed a generic method based on the
current approaches in landmarking to solve a variety
of problems that arose from the medical character of
the dataset:
• the method should be robust to missing teeth,
• tens of cases should be sufficient to train the net-
work,
• and the speed of the inference should be fast
enough to be used in a clinical application.
Especially valuable is the introduced post-
processing based on heatmap regression uncertainty
analysis and analysis of the uncertainty of the multi-
view approach. It ensures that our method cor-
rectly detects landmark presence without any addi-
tional computations. This is inevitable for orthodontic
flow as it robustly detects teeth presence even in chal-
lenging cases (e.g., already discussed 3rd molars).
This aspect was not discussed in recent works that
deal with orthodontics landmarks on teeth crowns.
In addition to the post-processing and the method
itself, this paper presents valuable comparisons and
experiments on various factors that impact the effi-
ciency of alternative variations of the method:
• rendering type of the processed 3D object to be
used as an input (depth map, geometry or combi-
nation of both),
• comparison of several network designs (U-Net,
Attention U-Net, and Nested U-Net),
• the analysis of the results of two consensus meth-
ods: a method that calculates the centroid of mul-
tiple predictions and a geometric method based on
the RANSAC algorithm and least-squares fit,
• and the analysis of the correlation between the
number of viewpoints and the method accuracy.
3 DATASET OF 3D DENTAL
SCANS AND LANDMARKS IN
THIS STUDY
Our method was trained and evaluated on a dataset of
337 3D dental scans of human dentition represented
as polygon meshes. The dataset contains cases of
both maxillary and mandibular dentition. Since all
dentition scans were anonymized, it is not possible to
undertake complex analysis of patients’ age or ethnic-
ity. Therefore, the data analysis was empirical and fo-
cused on aspects such as the frequency of absence of
teeth, the rate of healthy dentition, and dentition with
malocclusion and shifted teeth. Concerning these as-
pects, our data reflect real orthodontics patients since
BIOIMAGING 2022 - 9th International Conference on Bioimaging
26
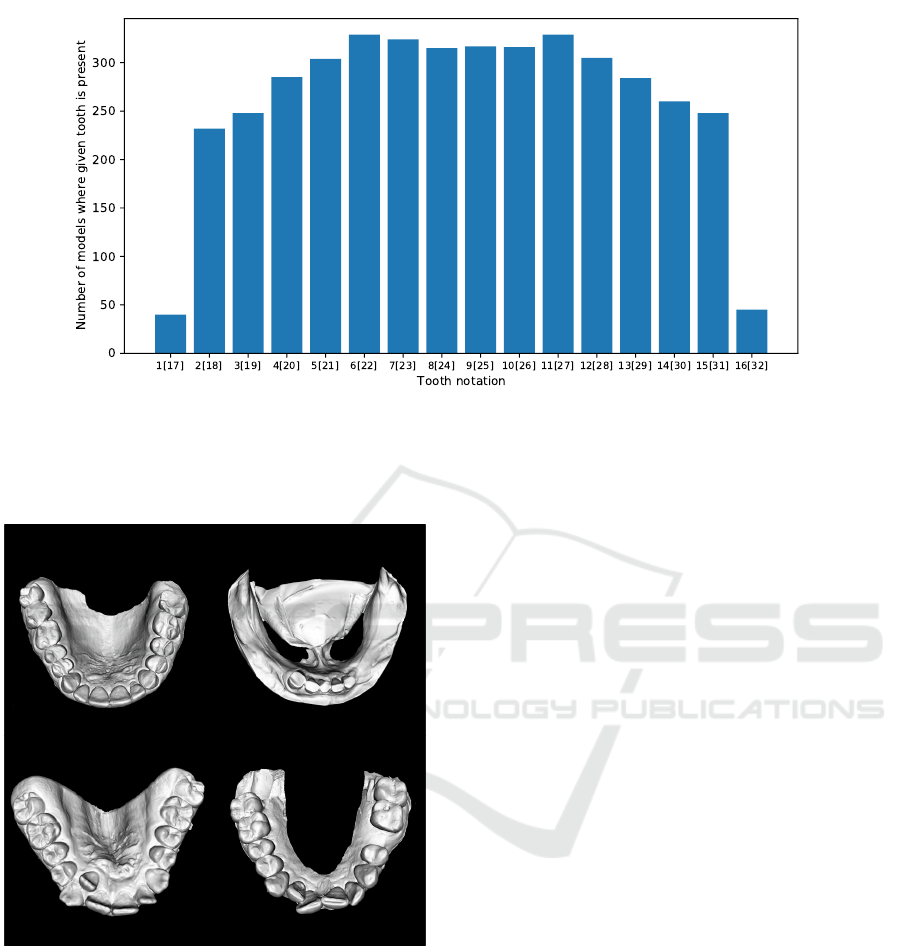
Figure 2: Distribution of casts where given tooth is present on the dentition. For example, out of 337 scanned dentition from
the dataset, less than 50 cases contain either left or right 3rd molar. This distribution reflects the reality, as 3rd molars are
often extracted (Normando, 2015). On the other hand, canines and incisors are present in the vast majority of models. Please
note that the Universal Numbering System is used to refer teeth. Also note that teeth 1 and 17 are considered as the same
category, likewise to the rest of the teeth.
Figure 3: Examples of dental casts within the dataset. Data
were collected from orthodontics patients, so patients usu-
ally suffer from different kinds of malocclusion, as depicted
on the bottom examples.
the diversity of data is significant, which is essential
for the algorithm’s robustness. Figure 3 depicts the
variety of dentitions in our dataset. The frequency
of missing teeth confirms the diversity in orthodon-
tics cases as well. Figure 2 shows the number of
cases where individual teeth are not missing within
the dataset. Landmarks used in this study address the
digital orthodontics flow in the existing planning soft-
ware. These landmarks define the mesial and distal
location of each tooth. They are placed on the oc-
clusal surface of molars and premolars and the incisal
surface on canines and incisors, as close to the cheek-
facing surfaces as possible. In other words, 32 land-
marks must be placed on one arch in case of full den-
tition, two for each tooth. Ground truth positions of
landmarks were annotated by one person only.
4 PROPOSED SOLUTION FOR
ORTHODONTICS LANDMARK
DETECTION
An outline of our method can be found in Figure 4.
Prior to each evaluation, there is a precondition to
align the evaluated mesh so the occlusal surfaces face
the camera. Afterward, following the multi-view ap-
proach, the model is observed from various camera
extrinsics. We used uniformly distributed positions
of the camera with a maximal angle of ± 30 degrees
from the initial aligned position.
Network Inputs and Outputs
Images in the form of depth maps and direct rendering
of the geometry are used as the inputs to the neural
network.
From each acquired view, features are extracted
and processed in the heatmap regression manner. In
a similar way as in (Pfister et al., 2015), during train-
ing, the input example is denoted as a tuple (X, y),
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking
27
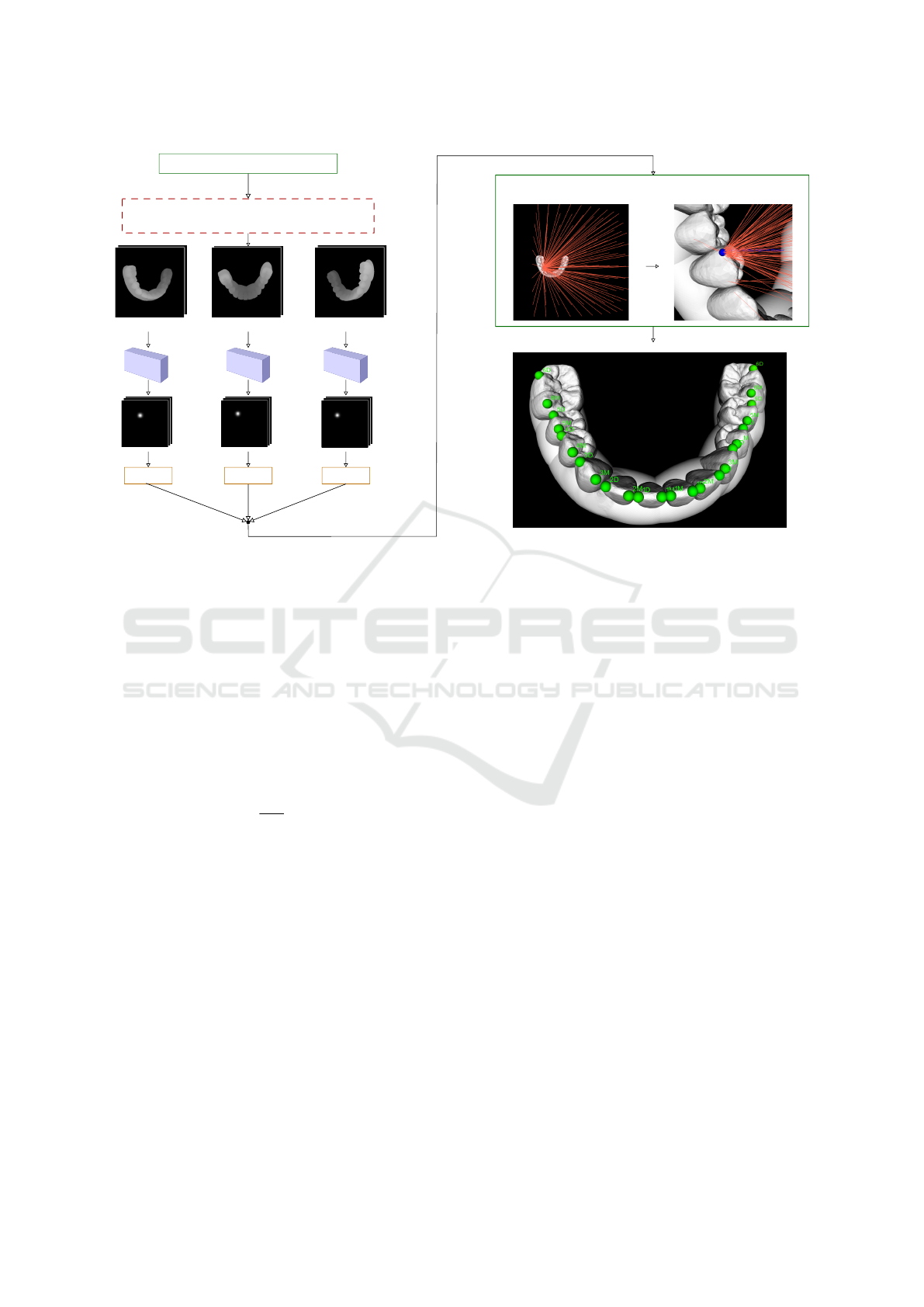
Multi-view rendering
3D scan in STL format
View 1
... View 1 < n < N ... View N
CNN
CNN
CNN
... ...
... ...
...
}
32
...
}
32
...
}
32
... ...
2D ↦ 3D
2D ↦ 3D
2D ↦ 3D
Predictions: points/rays
Consensus method
+
Octree based-search division
Result: predicted landmarks in 3-space
For each landmark
}
Depth map
+ geometry
... ...
Figure 4: Outline of the proposed method for orthodontics landmark detection. Following the multi-view approach, input 3D
model is observed from various viewports and sent to the CNNs to produce heatmaps. Landmark screen coordinates are ex-
tracted from obtained heatmaps and further processed by the consensus method, which produces final estimates. Additionally,
the maximum value in the activation map, together with the output of the consensus method, are used to detect tooth presence
during post-processing.
where X is the 2-channel input and y stands for the
coordinates of 32 landmarks located in input X. Fur-
thermore, the training data are denoted as N = {X, y}
and the network regressor as φ. Then, the training ob-
jective is the estimation of the network weights λ:
argmin
λ
∑
(X,y)∈N
∑
i, j,k
kG
i, j,k
(y
k
) − φ
i, j,k
(X,λ)k
2
(1)
where G
i, j,k
(y
i
) =
1
2πσ
2
e
−[(y
1
k
−i)
2
+(y
2
k
− j)
2
]/2σ
2
is
a Gaussian centered at landmark y
k
with fixed σ.
Using this approach, the last convolutional layer’s
output is a heatmap represented as a fixed-size
i × j × 32-dimensional matrix. This implies that the
the predicted results are 32 channels (as we intend to
predict 32 landmarks in our data).
Interpretation of Heatmap Regression Output in
Terms of 3D Data
The predicted 2D heatmap can be interpreted as the
landmark’s screen coordinate (in IR
2
) position (x, y).
Each output channel contains a heatmap with a Gaus-
sian representing the probability of a given land-
mark’s screen coordinate in each pixel. Thus, the re-
sulting screen coordinate must be extracted from the
predicted heatmap by finding coordinates of the peak
value. It is indispensable to propagate the coordinates
into a world coordinate system IR
3
and find a final es-
timate by combining outputs from all camera views.
With the known position of the center of projec-
tion, the prediction for a single view of one landmark
can be interpreted as (i) a ray defined by the origin in
the corresponding center of projection and the point
on the view plane at detected screen coordinates or
(ii) simply a point in the 3D scene, i.e. the converted
display coordinate into 3D space.
Consensus Methods
These individual predictions are combined in a con-
sensus method, which is a standard post-processing
step in the multi-view approach. Based on the maxi-
mum value in the activation map, only certain predic-
tions above the experimentally determined threshold
are sent to the consensus method. Certainty analy-
sis will be discussed later in this work. If the pre-
dictions are interpreted as rays, the consensus method
combines the RANSAC algorithm to eliminate partial
predictions classified as outliers with the least-squares
fit.
To achieve this, we defined each ray by its origin
a
i
and a unit direction vector n
i
, similarly as (Paulsen
et al., 2018). Then, the sum of squared distances from
BIOIMAGING 2022 - 9th International Conference on Bioimaging
28

a point p is calculated as follows:
∑
i
d
2
i
=
∑
i
[(p − a
i
)
T
(p − a
i
) − [(p − a
i
)
T
n
i
]
2
]. (2)
It is necessary to differentiate this equation with re-
spect to p. It brings the solution p = S
+
C, where
S
+
denotes the pseudo-inverse of S. In this case,
S =
∑
i
(n
i
n
T
i
− I) and C =
∑
i
(n
i
n
T
i
− I)a
i
.
The RANSAC procedure initially estimates the value
of p by three randomly chosen rays. The residual is
computed as the sum of squared distances (see Equa-
tion 2) from p to the included rays, and the iterative
RANSAC algorithm then performs I iterations. In
each of these iterations, the number of inliers and out-
liers is calculated, respecting a predefined threshold τ.
This is a minimizing task that finds a point in IR
3
with
the shortest distance to all remaining lines.
This method can be interchanged with a more sta-
tistical approach that is less computationally demand-
ing, and it simply finds the mean position of the pre-
dicted points. Let’s consider N as the number of views
used in the multi-view approach. Let’s also interpret
the single-view evaluation output as a point on the tar-
get polygonal model. With N views, the final out-
put P is a single point in IR
3
and is calculated from
N points as a mean value of these points.
Finding Closest Point on Mesh Surface
These methods find the estimation among multiple
predictions, but do not guarantee that the predicted
landmark is placed on the surface of the evaluated
polygonal model. Thus, the last necessary step is to
find the closest point on the surface of the polygonal
model. An octree data structure contains a recursively
subdivided target polygonal model. The center of the
closest face on the surface of the polygonal model to
the consensus output is considered the final estimate.
4.1 Post-processing for Classification of
Teeth Presence
As discussed in previous sections, assuming that the
evaluated 3D scan represents full dentition would be
loose. Therefore, apart from the accurate placement
of the present landmarks, our post-processing con-
tains an analysis of the presence of each tooth (i. e., of
corresponding couple of landmarks). This is in fact
a binary classification task, whose result is based on
two uncertainty hypotheses:
• Like in (Drevick
´
y and Kodym, 2020), the net-
work is trained to regress heatmaps with the am-
plitude of 1. Then, the fundamental assumption
0
50
100
0
20
40
60
80
100
120
Peak value: 0.012, GT Presence: False
0.000
0.002
0.004
0.006
0.008
0.010
0.012
0
50
100
0
20
40
60
80
100
120
Peak value: 0.7838, GT Presence: True
0.0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
Figure 5: Examples of predicted heatmaps and analysis of
the uncertainty. The top picture illustrates an example of
a prediction with low peak value (0.012). Referencing to
corresponding ground truth, this landmark is not present on
the surface of the polygonal model. The bottom picture, on
the other hand, shows the opposite situation. According to
the ground truth, the peak value is relatively high, and this
landmark is really present on evaluated polygon mesh. Note
that the maximal amplitude value in a heatmap is 1.
is that during the inference, the certainty is mea-
sured by the maximum value in the activation
map, with a proportional increase to the network’s
confidence (Maximum Heatmap Activation Confi-
dence). See Figure 5 for an example.
• The RANSAC consensus method robustly esti-
mates the landmark position by eliminating out-
lier predictions. Thus, the proportion of inliers
and outliers is another valuable output of this con-
sensus method, assuming the number of inliers is
proportional to the overall confidence (Multi-view
Confidence).
These assumptions result in a threshold value,
which combines the Maximum Heatmap Activation
Confidence and Multi-view Confidence, both in a unit
range and equally weighed. The optimal threshold
value can be determined by standard approaches for
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking
29

a binary classifier, as an example by the ROC curve.
This goes to show that such post-processing delivers
vital data for classification of landmarks presence by
self-evaluation, i. e., no additional computations or
network evaluations are needed to obtain such infor-
mation. Having the requirement of low computational
time in mind, this is more than eligible.
5 EXPERIMENTS AND RESULTS
To find the best possible results, we experimen-
tally investigated and compared several parts of the
method:
• Architecture Design: we compared the U-Net ar-
chitecture with two of its offshoots: the Attention
U-Net and the Nested U-Net.
• Consensus Methods: a comparison of RANSAC
consensus method with centroid calculation is
presented.
• Viewpoint Numbers for the Multi-view Ap-
proach: we analysed whether the increase of
viewpoint number has an impact on the method
accuracy. We experimented with 1, 9, 25, and
100 views.
• CNN Inputs: depth map, direct geometry ren-
dering and its combination (2-channel input) were
compared.
All metrics are measured in physical units (mm)
since the end clinical application is related to physical
units.
5.1 Training Procedure
The input to the neural network is either a single-
channel depth map, single-channel image of the ren-
dered geometry, or two-channel combination of both,
depending on experiment. In all cases, the size of
input was set to 128 × 128. The training procedure
ran on an NVIDIA GeForce RTX 2060 with 6 GB of
memory.
The dataset of 337 dental scans was divided into
a set of 247 cases used for training and a test set of
90 cases. Furthermore, the training set was split in
the ratio of approximately 4:1 into a training and val-
idation set, respectively.
Following augmentation techniques were applied
to both, the 2D input(s) and the ground truth
heatmaps:
• Scale: in the range [0.90, 1.10],
• Rotation: in the range [−30, 30] degrees,
• Translation: in the range [−10 px, 10 px] and ap-
plied in both vertical and horizontal directions.
Training Parameters and Loss Function
Networks were trained using the Adam optimizer
with the weight decay set to 10
−3
. The learning rate
was initially set to 10
−3
. Its value was dynamically
reduced using learning rate scheduler. The learning
rate was reduced by a factor of 0.5 every time the
value of validation loss has not improved for 5 con-
secutive epochs. The validation loss was monitored
for the early stopping. If the validation loss value
did not improve for more than 20 consecutive epochs,
the training was stopped. To reduce the memory re-
quirements during training, the automatic mixed pre-
cision was used. The batch size was set to 32. To
train the models on a regression problem, the Root
Mean Square Error (RMSE) loss was used.
5.2 Overall Results
The main focus of the experiments was to find the
best setup of the method. Overall results are sum-
marized in Table 1. Our results show that the ac-
quired accuracy is mostly influenced by the consensus
method, where RANSAC outperforms the Centroid
by a large margin in all setups. As for the used ar-
chitecture, the overall results show that the Attention
U-Net performs slightly better than the rest. Combi-
nation of depth maps and geometry renders impacts
the results in a positive way as well. See Figure 7 for
box plots of radial errors of individual detected land-
marks. The Attention U-Net has 526 534 trainable pa-
rameters and inference takes 4 seconds on average on
Intel Core i7-8750H CPU @ 2.20 GHz with 6 cores
(using 25 views).
When comparing our results to the framework
from (Wu et al., 2021), specifically with their best-
performing strategy, 2-stage iMeshSegNet+PointNet-
Reg. In terms of accuracy, they achieve a slightly
better error of 0.623 ± 0.718 mm. Their approach
slightly outperforms ours (in best-performing con-
figuration, 0.75 ± 0.96 mm), but it is necessary
to keep in mind several factors. As a matter of
fact, their dataset consists of 36 samples. Such rel-
atively small number should be increased to ensure
the method’s robustness to the large variability of or-
thodontic cases. Our dataset is more challenging and
consists of problematic cases with severe teeth shift-
ings and of many cases with missing teeth. In addi-
tion, they detect landmarks only on 10 teeth, exclud-
ing, for example, very problematic 3rd molars. Thus,
for a fair comparison, it would be vital to benchmark
BIOIMAGING 2022 - 9th International Conference on Bioimaging
30

Table 1: Overall results of the individual networks with different multi-view settings. Table compares the system performance
with different combinations of architectures, network inputs, consensus methods, and number of viewpoints. A combination
of the Attention U-Net architecture, the RANSAC consensus method, and 100 rendered views achieves the best performance.
R stands for the mean radial error, and SD stands for standard deviation. Values are calculated from all predicted landmarks on
dental scans from the test dataset and measured in millimeters (mm). All values are measured on networks with class-balanced
loss. Please note that the alignment of evaluated 3D scans influence the measured values.
Single-view
Multi-view
Architecture & N = 9 N = 25 N = 100
consensus method R SD R SD R SD R SD
BN U-Net (Depth)
Centroid
2.24 4.02
2.00 2.37 1.74 2.33 1.80 1.96
RANSAC 1.24 2.86 1.02 3.75 1.01 4.28
BN U-Net (Geom)
Centroid
2.13 4.41
2.03 3.14 1.69 2.21 1.67 2.41
RANSAC 1.20 3.01 1.17 2.16 1.06 2.22
BN U-Net (Depth & Geom)
Centroid
2.02 4.10
1.90 2.12 1.82 2.48 1.85 3.23
RANSAC 1.01 3.77 0.84 2.05 0.77 1.94
Att U-Net (Depth)
Centroid
1.73 3.48
2.37 3.37 2.02 2.87 2.01 1.99
RANSAC 1.18 1.88 1.10 2.05 0.95 1.62
Att U-Net (Geom)
Centroid
1.72 3.62
2.31 2.68 1.98 2.09 1.96 2.38
RANSAC 1.14 1.51 1.02 3.75 0.91 1.11
Att U-Net (Depth & Geom)
Centroid
1.67 3.06
2.00 2.37 1.74 2.33 1.80 1.96
RANSAC 0.93 1.03 0.79 1.01 0.75 0.96
Nes U-Net (Depth)
Centroid
1.77 3.32
2.29 2.12 2.32 1.99 2.12 3.04
RANSAC 1.09 2.60 1.00 1.85 0.95 2.82
Nes U-Net (Geom)
Centroid
1.77 3.00
2.44 1.98 2.30 3.01 2.23 2.58
RANSAC 1.11 1.83 0.93 1.67 0.93 1.99
Nes U-Net (Depth & Geom)
Centroid
1.69 2.62
2.30 3.18 2.31 2.72 2.16 2.55
RANSAC 0.98 2.09 0.83 2.12 0.80 1.45
our results on a public dataset, which is not currently
available.
Impact of Viewpoint Number
As for the number of views used in the multi-view ap-
proach, a negligible increase in accuracy is achieved,
comparing 25 and 100 views. This increase in view-
point number, however, significantly raises the infer-
ence time, so it is necessary to cross-validate this
number to obtain desirable accuracy as well as com-
putational time. For example, an increase of 0.04 mm
in accuracy as a trade-off for 4× higher inference time
is considerable. See Figure 6, which analyzes the
Success Detection Rate (SDR) of various numbers of
views.
Robustness to Model Rotations
Generally speaking, the multi-view approach is not
invariant to rotation. The requirement of initial model
alignment stems from this matter of fact. Therefore,
we were interested in how the method performs with
increasing alignment error. With an alignment error
of less than 20 degrees, the method brings sufficiently
accurate predictions. With higher alignment errors,
especially above 30 degrees, the results should be vi-
sually checked and if needed, manually fixed. This
correlation is depicted in Figure 8.
2.0
2.5
4.0
Acceptance value (mm)
75
80
85
90
95
100
SDR (%)
78.32
93.36
97.98
98.07
1 view
9 views
25 views
100 views
Figure 6: Success Detection Rates (SDRs) for Attention U-
Net, 2-channel input and the RANSAC consensus method.
Assuming the acceptable distance is 2 mm, setting the num-
ber of viewpoints higher than 25 does not bring any signifi-
cant increase in performance.
5.3 Detection of Teeth Presence
The main focus of the experiments was to determine
whether the method’s self-evaluation can detect the
presence of landmarks (and therefore, teeth). In line
with previous studies in uncertainty measures, each
prediction’s peak value is considered one of the de-
cision factors. Networks were trained by regressing
heatmaps containing a Gaussian activation with the
amplitude of 1. The predictions should follow the
similar trend. There was no Gaussian in the ground
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking
31
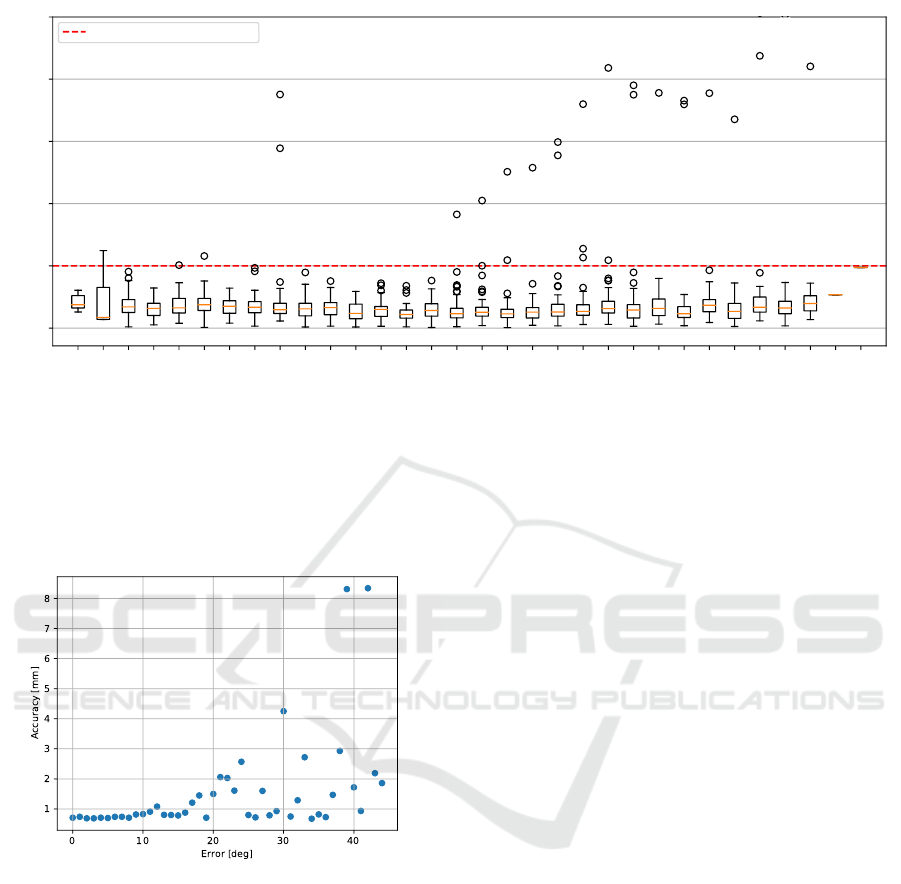
L8D
L8M
L7D
L7M
L6D
L6M
L5D
L5M
L4D
L4M
L3D
L3M
L2D
L2M
L1D
L1M
R1M
R1D
R2M
R2D
R3M
R3D
R4M
R4D
R5M
R5D
R6M
R6D
R7M
R7D
R8M
R8D
Landmark
0
2
4
6
8
10
Distance from GT (mm)
Acceptable distance (2 mm)
Figure 7: Box plots of the radial error values of individual landmarks. These values were measured with following method
configuration: Attention U-Net, two-channel input, 25 views, and RANSAC consensus method. Additionally, the class-
balanced loss was used for training. The landmark notation describes the type of landmark as follows: L stands for Left
dentition part and R for Right, values 1 - 8 describe tooth in the quadrant (1 for centran incisor and 8 for 3rd molars) and
letters M and D stand for mesial and distal landmark, respectively. Note that the outlier values in Right dentition part were
caused by one problematic case, where all teeth in right part were shifted by one and our method misclassified each tooth with
its adjacent tooth.
Figure 8: Correlation between error from required align-
ment and landmarking accuracy. As the 3D model is ob-
served from different angles, the method robustly estimates
landmarks even when the model is slightly rotated. Over-
all, the method becomes less stable with increasing error in
alignment, especially above 30 degrees.
truth image if a landmark was missing on the polyg-
onal model during training. This implies that the pre-
dictions should be either heatmaps with a peak value
close to 1 or heatmaps with all values close to 0.
By plotting an ROC curve, it was found that the
threshold value that brings off the best sensitivity and
specificity values is 0.375. Please note that this value
should be always cross-validated for each task. The
accuracy of the detection was 96.36%. After em-
pirical observations, there were situations where on
a tooth, one landmark was classified as missing and
the second one as present. This undesirable situa-
tion was eliminated by measuring the certainty in cou-
ples, averaging its confidences. It leads to better re-
sults, even if the improvement is negligible, achiev-
ing an accuracy of 96.69%. Another promising find-
ing comes from the RANSAC consensus method out-
put. The Multi-view Confidence, measured as the
ratio between inliers and outliers, was again moni-
tored by an ROC curve. The threshold was set to
0.85 and combined with the analysis of heatmap max-
imum value. Superior results are seen for this combi-
nation, as 97.68% of landmarks are correctly classi-
fied as missing or present.
Detecting Presence of 3rd Molars
A special category of detected teeth is 3rd molars. As
discussed in Section 3, those teeth are represented in
approximately 15% of the cases. The approach uti-
lized for detection of teeth presence suffers from this
imbalance, as the 3rd molars were always classified as
missing. This was due to the training, where, in most
cases, wisdom teeth were not present. To address this
problem, the loss was balanced in class-wise man-
ner (Cui et al., 2019). With this technique, 9 out of 12
wisdom teeth in the test set were correctly detected.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
32

0.0
mm
2.0 mm
4.0 mm
6.0 mm
8.0 mm
10.0+ m
m
Figure 9: Examples of automatically detected landmarks
with our method. Majority of predictions have the landmark
localization error less than 2 mm. Our method correctly de-
tects if a tooth is missing and does not produce predictions
of corresponding landmarks.
6 CONCLUSIONS
The present findings confirm that the multi-view
approach combined with the RANSAC consensus
method brings promising results in the automation of
landmark detection. Evaluated on a dataset of real or-
thodontics dental casts with significant diversity, the
method performs the best with Attention U-Net archi-
tecture and with two-channeled input of depth maps
and geometry renders. This method setup achieves
a landmarking accuracy of 0.75 ± 0.96 mm.
Importantly, we have also shown that the uncer-
tainty measures based on the analysis of the max-
imum values of regressed heatmap predictions in
combination with multi-view uncertainty yield con-
venient information in the process of landmark pres-
ence detection. Combining these uncertainty mea-
sures, our method correctly detects landmark pres-
ence in 97.68% of cases. This means that the method
is suitable to be applied to data where landmarks’
presence is not granted. In addition, the method meets
the needs of clinical applications, as the inference at
the user’s side takes seconds to be calculated, even on
less powerful CPUs.
Even though the accuracies are satisfying, the size
of the dataset could not cover every bit of a maloc-
clusion case and teeth shifting. Future research could
examine the method on a larger dataset of dentition
with even more complex cases. Furthermore, future
studies should focus on the improvements in the in-
variance of rotation. The association between the ro-
tation from the aligned position and the landmarking
accuracy was investigated in this work, and it is the
main shortcoming of the proposed method.
ACKNOWLEDGEMENTS
This work was supported by TESCAN 3DIM, s.r.o.,
which provided us with the dataset used in this work
as well as with its funding.
REFERENCES
Cui, Y., Jia, M., Lin, T.-Y., Song, Y., and Belongie, S.
(2019). Class-balanced loss based on effective num-
ber of samples.
Donner, R., Menze, B. H., Bischof, H., and Langs, G.
(2013). Global localization of 3d anatomical struc-
tures by pre-filtered hough forests and discrete opti-
mization. Medical Image Analysis, 17(8):1304–1314.
Drevick
´
y, D. and Kodym, O. (2020). Evaluating deep learn-
ing uncertainty measures in cephalometric landmark
localization. In Proceedings of the 13th International
Joint Conference on Biomedical Engineering Systems
and Technologies - BIOIMAGING,, pages 213–220.
INSTICC, SciTePress.
Fischler, M. A. and Bolles, R. C. (1981). Random sample
consensus: A paradigm for model fitting with appli-
cations to image analysis and automated cartography.
Commun. ACM, 24(6):381–395.
Lv, J., Shao, X., Xing, J., Cheng, C., and Zhou, X.
(2017). A deep regression architecture with two-stage
re-initialization for high performance facial landmark
detection. In 2017 IEEE Conference on Computer Vi-
sion and Pattern Recognition, CVPR 2017, Honolulu,
HI, USA, July 21-26, 2017, pages 3691–3700. IEEE
Computer Society.
Normando, D. (2015). Third molars: To extract or not to ex-
tract? Dental press journal of orthodontics, 20(4):17–
18.
Paulsen, R. R., Juhl, K. A., Haspang, T. M., Hansen, T. F.,
Ganz, M., and Einarsson, G. (2018). Multi-view con-
sensus CNN for 3d facial landmark placement. In
Jawahar, C. V., Li, H., Mori, G., and Schindler, K.,
editors, Computer Vision - ACCV 2018 - 14th Asian
Conference on Computer Vision, Perth, Australia, De-
cember 2-6, 2018, Revised Selected Papers, Part I,
Robust Teeth Detection in 3D Dental Scans by Automated Multi-view Landmarking
33

volume 11361 of Lecture Notes in Computer Science,
pages 706–719, Cham. Springer.
Payer, C., Stern, D., Bischof, H., and Urschler, M. (2016).
Regressing heatmaps for multiple landmark localiza-
tion using cnns. In Ourselin, S., Joskowicz, L.,
Sabuncu, M. R.,
¨
Unal, G. B., and Wells, W., editors,
Medical Image Computing and Computer-Assisted In-
tervention - MICCAI 2016 - 19th International Con-
ference, Athens, Greece, October 17-21, 2016, Pro-
ceedings, Part II, volume 9901 of Lecture Notes in
Computer Science, pages 230–238. Springer.
Pfister, T., Charles, J., and Zisserman, A. (2015). Flowing
convnets for human pose estimation in videos. In 2015
IEEE International Conference on Computer Vision,
ICCV 2015, Santiago, Chile, December 7-13, 2015,
pages 1913–1921. IEEE Computer Society.
Qi, C. R., Su, H., Mo, K., and Guibas, L. J. (2017). Pointnet:
Deep learning on point sets for 3d classification and
segmentation.
Su, H., Maji, S., Kalogerakis, E., and Learned-Miller, E. G.
(2015). Multi-view convolutional neural networks for
3d shape recognition. In 2015 IEEE International
Conference on Computer Vision, ICCV 2015, Santi-
ago, Chile, December 7-13, 2015, pages 945–953.
IEEE Computer Society.
Sun, D., Pei, Y., Li, P., Song, G., Guo, Y., Zha, H., and Xu,
T. (2020). Automatic tooth segmentation and dense
correspondence of 3d dental model. In MICCAI.
Sun, Y., Wang, X., and Tang, X. (2013). Deep convolutional
network cascade for facial point detection. In 2013
IEEE Conference on Computer Vision and Pattern
Recognition, Portland, OR, USA, June 23-28, 2013,
pages 3476–3483. IEEE Computer Society.
Wu, T.-H., Lian, C., Lee, S., Pastewait, M., Piers, C., Liu, J.,
Wang, F., Wang, L., Jackson, C., Chao, W.-L., Shen,
D., and Ko, C.-C. (2021). Two-stage mesh deep learn-
ing for automated tooth segmentation and landmark
localization on 3d intraoral scans.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
34
