
Identification of Planarian Individuals by Spot Patterns in Texture
Nikita Lomov
1 a
, Kharlampiy Tiras
2,3 b
and Leonid Mestetskiy
1,4 c
1
Federal Research Center “Computer Science and Control” of the Russian Academy of Sciences, Moscow, Russia
2
Institute of Theoretical and Experimental Biophysics of the Russian Academy of Sciences, Pushchino, Russia
3
Pushchino State Institute of Natural Science, Pushchino, Russia
4
Lomonosov Moscow State University, Moscow, Russia
Keywords:
Animal Identification, Planarian Flatworms, Skeleton, Fat Curve, Point Registration, Assignment Problem.
Abstract:
Planarian flatworms are known for their abilities to regenerate and are a popular biological model. Identi-
fication of individual planarian individuals is useful for automating biological research and improving the
accuracy of measurements in experiments. The article proposes a method for identifying planaria by their
texture profile, characterized by a set, shape, and position of light spots on the worm’s body— areas without
pigment. To make the comparison of planaria of different sizes and in different poses, the method of planarian
texture normalization is suggested. It is based on the selection of a main branch in the skeleton of a segmented
image and allows one to switch to a unified coordinate system. Also, a method for creating a generalized
textural profile of a planarian, based on averaging sets of spots for multiple images, is proposed. Experiments
were carried out to identify planaria for different types of observations—during one day, during several days
and during several days of regeneration after decapitation. Experiments show that light spots are a temporally
stable phenotypic trait.
1 INTRODUCTION
Freshwater flatworms planaria are one of those groups
of animals that are recognized as classical biologi-
cal models. The ability of adult planarians to mor-
phogenesis, that is, regeneration and asexual repro-
duction (Bagu
˜
n
`
a, 2012; Elliott and Alvarado, 2013;
Karami et al., 2015), is the most pronounced in the
animal kingdom. The only ones in the animal world,
planaria are even capable of regenerating their central
nervous system, the head ganglion, and this happens
in a very short time, from one to three weeks. The de-
velopment of digital technologies for the creation and
analysis of images has made it possible to develop a
quantitative description of the morphogenesis of pla-
naria in vivo (Tiras et al., 2015; Tiras et al., 2021).
Planarians are also one of the potentially promising
objects in the study of the cellular basis of immunity,
which in planarians proceeds by phagocytosis of food
by all planarian cells, except for nerve and germ cells
(Sheimann and Sakharova, 1974). Taking into ac-
count the current interest in various biological models
a
https://orcid.org/0000-0003-4286-1768
b
https://orcid.org/0000-0002-1853-8285
c
https://orcid.org/0000-0001-6387-167X
related to the problems of cellular immunity, planari-
ans can become one of the promising models for the
study of phagocytosis in vivo (Tiras et al., 2018; Peiris
et al., 2014; Apyari et al., 2021).
However, the widespread use of planaria for solv-
ing various fundamental and applied problems is hin-
dered by a number of unresolved objective problems,
one of which is the problem of identifying planarian
individuals in the course of an experiment. Thus, a
feature of the biology of the asexual race of planaria
Girardia tigrina is their preference for group habita-
tion during experiments. In addition, when seated
alone, planaria of this species tend to separate after 24
hours, which interferes with sufficiently long experi-
ments, therefore, during the experiment, such planari-
ans are kept in a group of 25-30 individuals in order to
limit asexual reproduction (Sheimann and Sakharova,
1974; Tiras et al., 2018). However, with group keep-
ing, it is impossible to distinguish planaria from each
other, which limits the possibility of assessing the in-
dividual characteristics of the course of certain physi-
ological processes over a number of days.
In this work, an attempt was made to identify pla-
naria based on the features of their body surface tex-
ture. The species name of the planaria, Girardia tig-
Lomov, N., Tiras, K. and Mestetskiy, L.
Identification of Planarian Individuals by Spot Patterns in Texture.
DOI: 10.5220/0010802000003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
87-96
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
87

rina, is due to the patchy structure of their surface,
which is associated with the mosaic distribution of the
pigment epithelium. This circumstance opens up the
possibility of using the features of the body surface
texture of individual planarians as the basis for their
personal identification.
Such an opportunity will make it possible to as-
sess the dynamics of morphogenesis and phagocyto-
sis of planaria in vivo, which will open up the pos-
sibility of creating quantitative models of the course
of these biological processes. Such models, in turn,
will be useful in studying the possibilities of control-
ling complex biological processes at the level of the
whole organism.
2 RELATED WORK
The identification of individuals is an important task
for researchers in population ecology, ethology, bio-
geography and experimental biology. Although the
fundamental possibility of perfectly accurate identifi-
cation is provided by DNA analysis, as well as the use
of chipping in combination with GPS tracking, it is
more realistic to use cheaper and non-invasive meth-
ods, for example, based on photography and video
surveillance. The rapid development of computer
vision systems observed in the last decade and the
strengthening of interdisciplinary interaction between
fields of science expressed in diffusion of methods
and approaches give hope for significant progress in
this area. For example, popular methods of human
face recognition, pose estimation, and keypoint de-
tection, based on neural network models have been
successfully adapted to process individuals of higher
mammals of some species—brown bears (Clapham
et al., 2020), Amur tigers (Liu et al., 2019), chim-
panzees (Freytag et al., 2016).
A number of computer vision techniques using
hand-designed features have also been developed.
Depending on the particular species, the source of
signs can be stripes on the body for zebras (Lahiri
et al., 2011), dermal plates for turtles (Rao et al.,
2021), shape of the fin for sharks (Hughes and
Burghardt, 2017), etc. In these cases, the feature
description can be presented in the form of vari-
ous mathematical objects: graphs, trajectories, point
clouds.
Particular individuals of animals, simpler than
mammals, are usually recognized insufficiently dis-
tinguishable, and the problem of recognizing various
closely related species, for example, worms (Lu et al.,
2021), is more often considered. It is one of the types
of worms, planarian flatworms, that are object of our
(a) (b) (c)
(d) (e) (f)
Figure 1: Planarian individual captured within a short pe-
riod of time (a-c) and a stable fragment of its texture (d-f).
The sources of this fragment are marked in (a-c) with green
frames.
interest. Planaria are versatile and powerful model
system for molecular studies of regeneration, adult
stem cell regulation, aging, and behavior. The fol-
lowing features of the planaria can be noticed from
the point of view of the task of visual recognition.
1. As their very name refers to, planaria are almost
flat objects, and therefore their current appearance
is described with sufficient completeness by a sin-
gle two-dimensional image.
2. Planaria have a significant ability to bend, com-
press and stretch individual parts of the body, so
that their shape can be considered relatively rigid
only in the head region.
3. The coloration of planaria is very primitive and
does not allow the use of complex color charac-
teristics for the identification of individuals.
4. Planaria are very dependent on environmental
conditions: the properties of the solution in which
they live, the weather outside the window, the diet
and the nature of the food they eat.
5. The body of a planarian is translucent, so the ap-
pearance in photographs is highly dependent on
shooting conditions and lighting.
The influence of these factors is illustrated in Fig.
1 showing the same planarian, photographed at dif-
ferent times. The distribution of body width in these
instances is very different: in Fig. 1a the thickness
of the planarian changes relatively slightly from the
tail to the head, in Fig. 1b the planarian has a strong
thickening about halfway, in Fig. 1c—severe thicken-
ing in the neck-like region above the head. At the
same time, on the body of the planarian, there are
fragments of a characteristic texture, which are pre-
served from photograph to photograph—Fig. 1d-e.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
88

These are the same constellation of light spots—areas
marked by the absence of pigment. The idea of the al-
gorithm is to describe the texture of the planarian by
a set and location of the corresponding spots and to
organize the comparison procedure in such a way as
to include the search for mutual correspondence be-
tween spots in different photographs.
All these factors make the development of signif-
icant and stable feature of the appearance and shape
of planaria an urgent and challenging task. On the
other hand, like many other computer vision systems,
our approach will include several classical customary,
such as segmenting objects in an image, normalizing
their shape, and finding points of interest. They will
be discussed in the following sections.
3 OBJECT SEGMENTATION
(a) (b) (c)
(d) (e) (f)
Figure 2: Binarization stages.
Despite the fact that it is natural for planaria to live in
groups of 20-30 individuals, the quality of shooting
provided by the available microscope was not enough
to capture the entire group at once. Therefore, for
photographing, the planaria were moved one by one
into an intermediate vessel, and the segmentation task
was reduced to finding a single individual in the im-
age. Since most of the body of planaria is pigmented,
and they themselves live in a transparent solution, the
principle that planaria are darker than the background
(Fig. 2a) can be taken as the basis for binarization.
Nevertheless, it is necessary to take into account the
following conditions of shooting: firstly, due to the
distribution of light and optical effects, the brightness
of the image decreases towards its edges, and sec-
ondly, the walls of the vessel, which are also dark,
can get into the frame. Therefore, to normalize the
background, the morphological operation of opening
a grayscale image G with a disk of a fixed diameter,
exceeding the width of the planarian, was performed
(Fig. 2b). Then the original image was subtracted
from the background and the negative of the result
was taken (Fig. 2c).
Next, the image was binarized using the Otsu
method, the average intensity values for the object
µ
f g
=
∑
i j
g
i j
[g
i j
<t]
∑
i j
[g
i j
<t]
and background µ
bg
=
∑
i j
g
i j
[g
i j
≥t]
∑
i j
[g
i j
≥t]
and binarization is performed with the threshold t
0
=
µ
f g
+ 0.7(µ
bg
−µ
f g
).
After that, the connected component with the
largest area is selected as the object mask. However,
since the walls of the vessel can be similar to a pla-
narian in size, color and shape, an additional check is
carried out: the sought component cannot touch the
edges of the image during Otsu binarization of the
original (without background subtraction) image. For
example, in Fig. 2d the red component will be as-
signed to the vessel walls, and the blue one will be
assigned to the background as a result of checking the
image in Fig. 2e. Also, to eliminate noise at the bor-
der with the final component, an operation of mor-
phological opening is carried out, which gives us as a
result Fig. 2f.
4 MAIN AXIS EXTRACTION
Shape standardization is an important stage in solv-
ing problems related to the recognition of flexible ob-
jects. It can be briefly described as follows. Let there
be a segmented image B with an area D ⊂ R
2
related
to the object. It is required to find the transforma-
tion T : D → Ω, where Ω ∈ R
2
is a standard domain.
Usually, additional requirements are imposed on the
transformation, for example, injectivity or smooth-
ness. Sometimes, if it is known that the form itself
is segmented into several parts {D
i
}, then the require-
ment p ∈ D
i
→ T (p) ∈ Ω
i
should be satisfied. For
instance, this setting is used in (Qu and Peng, 2010)
for standardizing confocal images of fruit fly nervous
systems. As a rule, for standardization, a certain ref-
erence set is chosen, which is a curved axis of sym-
metry of an object or its part. So, this approach was
discussed in (Duyck et al., 2015) for straightening
species with strong bilateral symmetry such as mar-
bled salamanders, skinks and geckos. At the same
time, the situation with worms is relatively simple,
since their shape, in fact, appears to be the vicinity of
a single line.
Let this line be smooth and described by the
equation q(t) = (x(t),y(t)), t ∈ [0,l], and x
0
(t)
2
+
y
0
(t)
2
= 1. The tangent to this curve has the direction
u(t) = (x
0
(t),y
0
(t)), and the perpendicular is v(t) =
(y
0
(t),−x
0
(t)). Let there also exist d, s.t. ∀p ∈ D ∃t ∈
[0,l] : ||p −q(t)|| ≤ d, and d <
1
max
t∈[0,l]
√
x
00
(t)
2
+y
00
(t)
2
.
Identification of Planarian Individuals by Spot Patterns in Texture
89
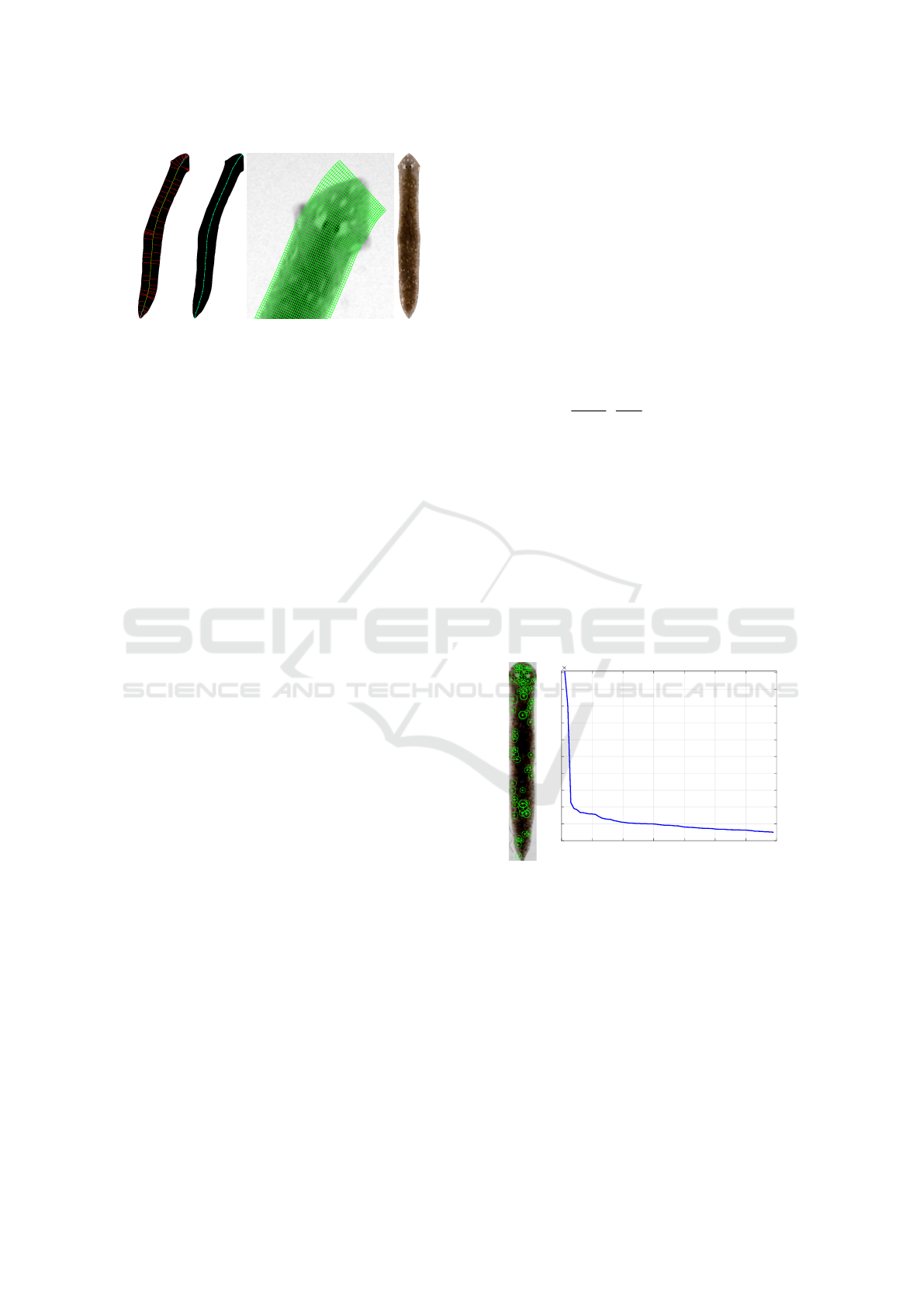
(a) (b) (c) (d)
Figure 3: Stages of main axis extraction.
Then the mapping
T
∗
(t,s) = (x(t)+(s−d)y
0
(t),y(t)−(s−d)x
0
(t)) (1)
is smooth, injective and maps the rectangle M =
[0,2d] ×[0, l] to the domain D
∗
⊃ D. Accordingly,
the transformation T = (T
∗
)
−1
maps D to the subset
of rectangle M, which, using scale transformations,
allows you to get a texture of a fixed size.
To determine the desired main axis of the worm,
in the work (Peng et al., 2007) an approach, based
on dividing the border into nominally left and right
parts, highlighting the middle line as a sequence of
midpoints of segments when traversing the left and
right parts and then refining the middle lines. Later
this algorithm was developed in (Flygare et al., 2013)
for better handling of worms of small eccentricity, i.e.
insufficiently elongated, with a shape close to elliptic.
To improve the algorithm, the coefficient of asymme-
try was determined between the left and right regions
of the worm’s body, into which the medial axis di-
vides the shapes. Note that since we are essentially
dealing with the search for an extended curvilinear
axis of symmetry of the worm, it is natural to use the
model of continuous skeleton (or, synonymously, me-
dial axis) of binary image (Mestetskiy and Semenov,
2008). The skeleton consists of lines equidistant from
two or more sections of the boundary (all solid lines
in Fig. 3a) and contains the required axis as a sub-
graph. Obviously, the shape of the worm should be
restored with sufficient completeness by the union of
the inscribed circles lying on the main axis—in fact,
we are talking about representing the worm’s shape
by a fat curve (Mestetskiy, 2000). Therefore, to se-
lect the base of the axis, the pruning of the skeleton
is performed—all branches of the skeleton that do not
make a significant contribution to the formation of the
shape are removed (green line in Fig. 3a). On the
other hand, the main axis should stretch from the top
of the head to the tip of the tail. These points are de-
fined as the points with the farthest projection on the
tangent rays to the ends of the base (dashed lines in
Fig. 3a), then the base is supplemented with paths in
the skeleton from the ends of the base to the found
points (blue lines in Fig. 3a).
To smooth the main axis, m points are sampled
evenly on it, which gives us a set of {x
k
,y
k
,r
k
}
m−1
k=0
,
where r
k
is the radius of the insribed circle centered
in (x
k
,y
k
). Then, using piecewise cubic Hermitian in-
terpolation (Fritsch and Carlson, 1980), we obtain the
smoothed version of the main axis: {x(t),y(t),r(t)},
t ∈ [0,m −1] (Fig. 3b). Let also the maximum ra-
dius of the inscribed circle on the curve be r
max
. We
set d = r
max
, so that the normalized texture fits into
the borders of the image. Then, for a texture of size
w ×h, a pixel with coordinates (i, j ) corresponds to
a point of the original image with the coordinates
T
(i, j) = T
∗
(
i(m−1)
h−1
,
j(2d)
w−1
), according to formula 1.
A fragment of the curvilinear grid is shown in Fig. 3c
and the straightened texture is shown in Fig. 3d. To
fill the texture, bilinear interpolation was used, and
pixels in positions (i, j) for which T
(i, j) /∈ D were
unmasked by alpha channel.
5 SEARCH FOR POINTS OF
INTEREST
5.1 Spot Extraction
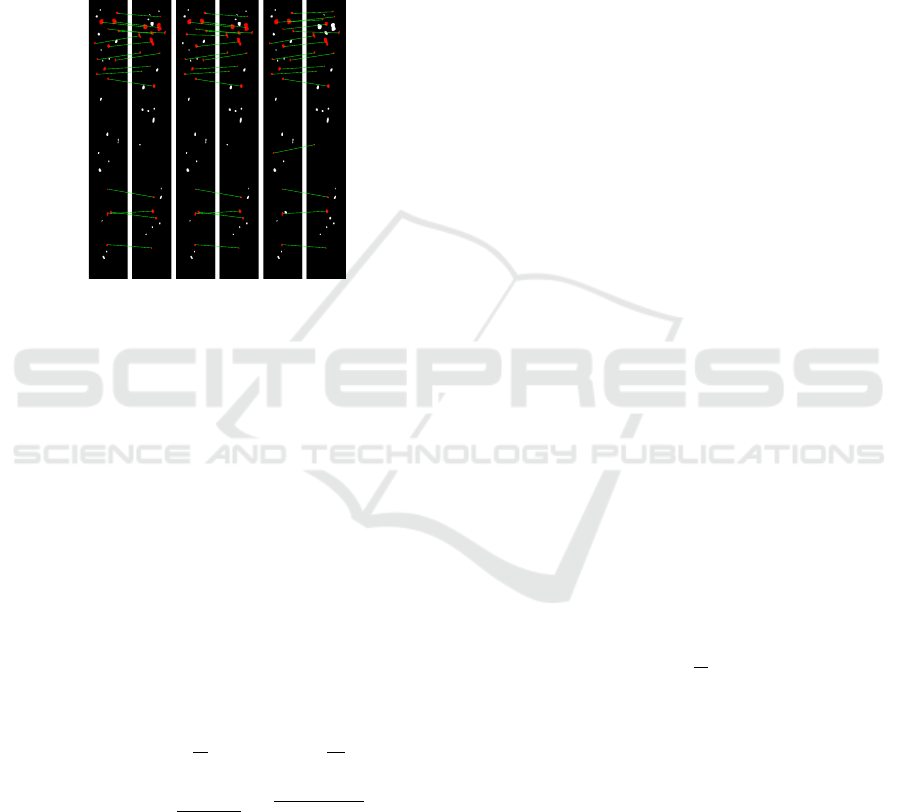
0 10 20 30 40 50 60 70
Rank
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
Strength
10
4
(a) (b)
Figure 4: SURF points with negative Laplacian value: lo-
cation (a) and strength sorted in descending order (b).
The surface of a planarian individual has a character-
istic pattern of light spots, that are the areas without
pigment. These spots are small in size and of prim-
itive, moreover, not always stable, shape. Although
one is able to match areas related to the same spots, in
different photographs of the same planaria, and also it
is intuitively clear what can be understood by “more
spotty” and “less spotty” planarians and parts of their
bodies, it is not possible to give an exact answer to the
question of how many spots are located on the body
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
90

Algorithm 1: Extraction of spots in texture.
Require: Grayscale image G, number of spots n, minimum
spot area s
min
, threshold step h
Ensure: Binary image B with spots
t = 1
B ← (G ∗LoG
σ
) ≤
1
t
m ← #{CC(B,s
min
)} m ← the number of conn.comp. of
B that are greater than s
min
if m = n then
Return
else if m < n then
while m < n do
t ←t + h
B ← (G ∗LoG
σ
) ≤
1
t
m ← #{CC(B,s
min
)}
end while
else
while m > n do
t ←t −h
B ← (G ∗LoG
σ
) ≤
1
t
m ← #{CC(B,s
min
)}
end while
B ← (G ∗LoG
σ
) ≤
1
t+h
end if
Leave in B n largest connected components
of a particular planarian. The situation is illustrated
by Fig. 4, where SURF points of interest (Bay et al.,
2008), which are lighter than the neighborhood, de-
tected in the texture, are shown, along with the plot
of their saliency depending on the rank. The graph
shows that, with the exception of a few first points, the
saliency changes very smoothly, which does not allow
setting an effective threshold rule due to a strong de-
crease in saliency. Moreover, the scalar products of
the feature vectors of these points are stably greater
than 0.5, and in more than half of the cases, exceed
0.9. This indicates that the main feature of spots is
their location, not their appearance, and also leads to
the idea of extracting a fixed number of spots from an
arbitrary texture. For this purpose, we use the Lapla-
cian Gaussian filter:
LoG
σ
(x,y) = −
1
πσ
2
1 −
x
2
+ y
2
2σ
2
exp
−
x
2
+ y
2
2σ
2
.
(2)
The spots will be defined as connective components in
the binary image B = (G ∗LoG
σ
) ≤t, with more than
half of the pixels belonging to the foreground. If the
image is too blurry, not enough contrast, or does not
contain a sufficient number of obvious spots, we will
use the linearity and stability (
∑
x
∑
y
LoG
σ
(x,y) = 0)
of the filter:
(aG + b) ∗LoG
σ
= a(G ∗LoG
σ
),
and we will select the binarization threshold instead
of increasing the contrast of the image itself. An al-
gorithm based on this principle is presented as Algo-
rithm 1.
5.2 Localization of Eyes
Light spots can occur in arbitrary places on the body
of a planarian, from head to tail. However, there are
two areas, which are also marked by the absence of
pigment and stand out as spots, but which are invari-
ably present in specific places—these are the eyes of
the planarian. The selection of eyes is important for
several purposes, the main one of which is the orien-
tation of the planarian texture (determining the head
end by the presence of eyes in it is a more reliable
criterion than, say, a wider head end as compared to
the tail end). We will determine the eyes as the best,
in some sense, pair of spots extracted at the previous
stage. Let (a
i
,x
i
,y
i
) be the area, abscissa and ordinate
of the center of the i-th spot found in the texture of
size w ×h. We use the following geometric properties
of the eyes:
• the area of the eye is usually larger than the area
of the regular spot;
• the eyes are located close to the border of the tex-
ture vertically;
• are located approximately at the same vertical
level;
• are located symmetrically about the vertical line
dividing the texture in half.
Texture n = 16 n = 24 n = 32 n = 40 n = 48
Figure 5: Extracting a different number of spots from a tex-
ture. Eye spots are highlighted in red.
For the spots with numbers (i, j), i 6= j we con-
struct the feature vector f
i j
containing the values
{a
i
,a
j
,min(y
i
,h −1 −y
1
),min(y
j
,h −1 −y
1
),y
i
−y
j
,
min(x
i
,w −1 −x
i
),min(x
j
,w −1 −x
j
),(x
i
+ x
j
−w + 1)}
and the monomes formed by them of order up to 3.
Then form a training sample from vectors of the form
f
kl
−f
i j
with label 1 and f
i j
−f
kl
with label −1, where
k and l refer to a pair of spots that are the eyes, and i
and j refer to the pair, marking no more than one eye.
Identification of Planarian Individuals by Spot Patterns in Texture
91
SVM with a linear kernel is used for classification and
when for the resulting model with weights w the pair
(t,s) maximizing hw,f
ts
i is taken as the eyes. The
results of the extraction of the spots and the selection
of the eyes are shown in Fig. 5.
6 COMPARISON OF SPOT
PATTERNS
(a) (b) (c)
Figure 6: Assignment problem solutions for (a) pure match-
ing (task 3) (b) matching with L
1
-term with α = 15 used
(task 4) (c) matching of ordered sets (rule 5 involved).
Due to differences in lighting, bending, and living
conditions of planaria, it is rarely possible to isolate
completely coincident sets of spots. Therefore, when
looking for a match one need to take into account that
part of the spots is the noise. In (Qu and Peng, 2010)
this problem arises when comparing skin marks in the
evaluation of soft biometrics and is solved by leaving
in the list of matches only top-50% of the best pairs.
It is also possible to draw an analogy with the point
registration task, which is to find the best match be-
tween two point clouds—fixed X and moving Y. In
one of the most popular methods for solving it, co-
herent point drift (Myronenko and Song, 2010), the
problem is reduced to maximizing the likelihood
n
∏
i=1
p(x
i
), p(x) = w
1
N
+ (1 −w)
M
∑
m=1
1
M
p(x|m),
where p(x|m) =
1
(2πσ
2
)
D/2
exp
−||x−T (y
m
,θ)||
2
2σ
2
, and
T (Y,θ) is a transformation T applied to
Y = (y
1
,...,y
M
) and parameterized by θ. The
method provides a soft assignment of points to the
components of a mixture of Gaussians with centers
in X, and the weight w of the uniform distribution
actually sets the proportion of noise in the data. We
will look for a strict match between the spots of
textures, but some of the points will not be match at
all.
For two textures characterized by the sets p = {p
i
}
and q = {q
j
}, we define the matrix C ∈ R
n×n
: c
i j
=
d(p
i
,q
j
), where d(p,q) is the dissimilarity measure
between spots. Let us define the dissimilarity between
sets as the minimum total dissimilarity between spots
in a one-to-one comparison, if only m ≤ n spots are
involved, which leads to the following task:
minimize
n
∑
i, j=1
c
i j
a
i j
subject to
n
∑
i=1
a
i j
≤ 1 for i = 1,...,n,
n
∑
j=1
a
i j
≤ 1 for j = 1,...,n,
n
∑
i, j=1
a
i j
= m,
a
i j
∈ {0,1} for i = 1 ≤ i, j ≤ n.
(3)
This formulation is a variation of the classical as-
signment problem, which can be solved by one of the
integer linear programming methods, for example, the
branch and cut method.
Let us pay attention to the fact that the situa-
tion when the residual vectors (x(p
i
) −x(q
j
),y(p
i
) −
y(q
j
)) between the matched spots are similar to each
other is much more natural than the strong scatter in
these vectors. For example, when the tail of a pla-
narian is squeezed, all spots should move downward
in relative texture coordinates. Thus, a term can be
added to the functional that takes into account the
total deviation from the mean of the residual vec-
tors. In order not to go beyond the scope of the
linear programming problem, we will act in the L
1
-
metric. Let X ,Y ∈ R
n×n
, x
i j
= x(p
i
) − x(q
j
), and
y
i j
= y(p
i
) −y
(
q
j
). Then, for example, the deviation
from the average along the Y -axis takes the form:
n
∑
i=1
n
∑
j=1
a
i j
y
i j
−
1
m
n
∑
i, j=1
a
i j
y
i j
.
Using the relaxing variables δ
i
,γ
i
∈R,i = 1,.. . , n,
the problem can be rewritten as:
minimize
n
∑
i, j=1
c
i j
a
i j
+ α
n
∑
s=1
δ
s
+ β
n
∑
t=1
γ
t
subject to
n
∑
i=1
a
i j
≤ 1 for i = 1,...,n,
n
∑
j=1
a
i j
≤ 1 for j = 1,...,n,
n
∑
i, j=1
a
i j
= m,
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
92

n
∑
i=1
a
i j
x
i j
−
1
m
n
∑
i, j=1
a
i j
x
i j
−δ
i
≤ 0 for i = 1,...,n,
n
∑
i=1
a
i j
y
i j
−
1
m
n
∑
i, j=1
a
i j
y
i j
−γ
i
≤ 0 for i = 1,...,n,
a
i j
∈ {0,1} for i = 1 ≤ i, j ≤ n,
δ
i
≥ 0,γ
i
≥ 0 for i = 1,...,n,
(4)
It can also be noted that, under possible defor-
mations of the planarian, the ordering of spots along
the Y -axis in the texture as a whole should be pre-
served. This property can be used in the formulation
of the functional, considering the ordered sets p and
q: i < j ⇒ y(p
i
) ≤ y(p
j
) & y(q
i
) ≤ y(q
j
) and adding
the following requirement to the 3 statement:
a
i j
= 1 & a
st
= 1 & i < s ⇒ j < t. (5)
The corresponding problem can be solved by dynamic
programming methods with recursive search for op-
timal matching k from l bottom spots. Visualiza-
tion of various methods of comparison for d(p,q) =
9(x(d) −x(q))
2
+ (y(d) −y(q))
2
is shown in Fig. 6.
7 GENERATION OF STANDARD
PATTERNS
Algorithm 2: Standard spot pattern calculation.
Require: Set of k sets of spots {p
i
= {p
i j
}
n
j=1
}
k
i=1
Ensure: Set of standard spots q = {q
i
}
n
i=1
Construct graph G = hV, Ei, where V is a set of all spots
in B and E is a set of all matches between spots in all
pairs in B
for i ← 1 to n do
V
0
← {v
0
}, v
0
= argmax
v∈V
#{e ∈ E : v ∈ e}
repeat
V
00
← {v ∈V \V
0
| ∃e = (v,v
0
) ∈ E : v
0
∈V
0
}
V
0
←V
0
∪V
00
until |V
0
| ≥ k or V
00
=
/
0
while |V
0
| > k do
v
00
← arg min
v∈V
0
#{e = (v,v
0
) ∈ E : v
0
∈V
0
}
V
0
←V
0
\{v
00
}
end while
for v ∈V
0
do
w(v) = #{e = (v, v
0
) ∈ E : v
0
∈V
0
}
f(v) is the feature vector of v
end for
q
i
←
∑
v∈V
0
w(v)f(v)
∑
v∈V
0
w(v)
V ←V \V
0
E ← E \{e ∈ E | e ∩V
0
6=
/
0}
end for
When solving the classification problem by searching
for the most similar texture among the standards, it is
necessary to carry out a comparison procedure with
each of the class samples in the database, which can
turn out to be a time-consuming task. Naturally, the
need arises to create a generalized set of spots that
would best characterize the entire class and would be
similar to all available samples at once. Obviously,
this set should consist of spots that are stably detected
from image to image, and such spots in different im-
ages will most likely be matched to each other. If we
represent the structure of correspondence of points in
textures in the form of a graph, then due to the absence
of connections between points of the same image, the
task will become similar to analyzing a k-partite graph
in order to find a clique (Dawande et al., 2001; Barber
et al., 2017) or a dense subgraph (Lee et al., 2010).
(a) (b) (c) (d) (e) (f) (g) (h) (i) (j)
Figure 7: Standard spot pattern generation. (a-h) Source
images, (i) locations of matched spots and (j) the resulting
standard. Some groups of matched spots are shown in spe-
cific colors.
To achieve a high speed of the procedure, a heuris-
tic algorithm of searching for a dense subgraph is pro-
posed based on expanding the neighborhood of a ver-
tex with a large number of connections and calculat-
ing the feature vector of a standard spot by averaging
the features of the vertices in the neighborhood. The
course of the procedure is described as Algorithm 2,
and its results are shown in Fig. 7.
8 EXPERIMENTS
8.1 Parameter Selection
The experiments were carried out with the following
parameters:
• number of points for main axis interpolation m =
64;
• texture height h = 1024;
• texture width w = 144;
• Gaussian filter deviation σ = 5;
Identification of Planarian Individuals by Spot Patterns in Texture
93
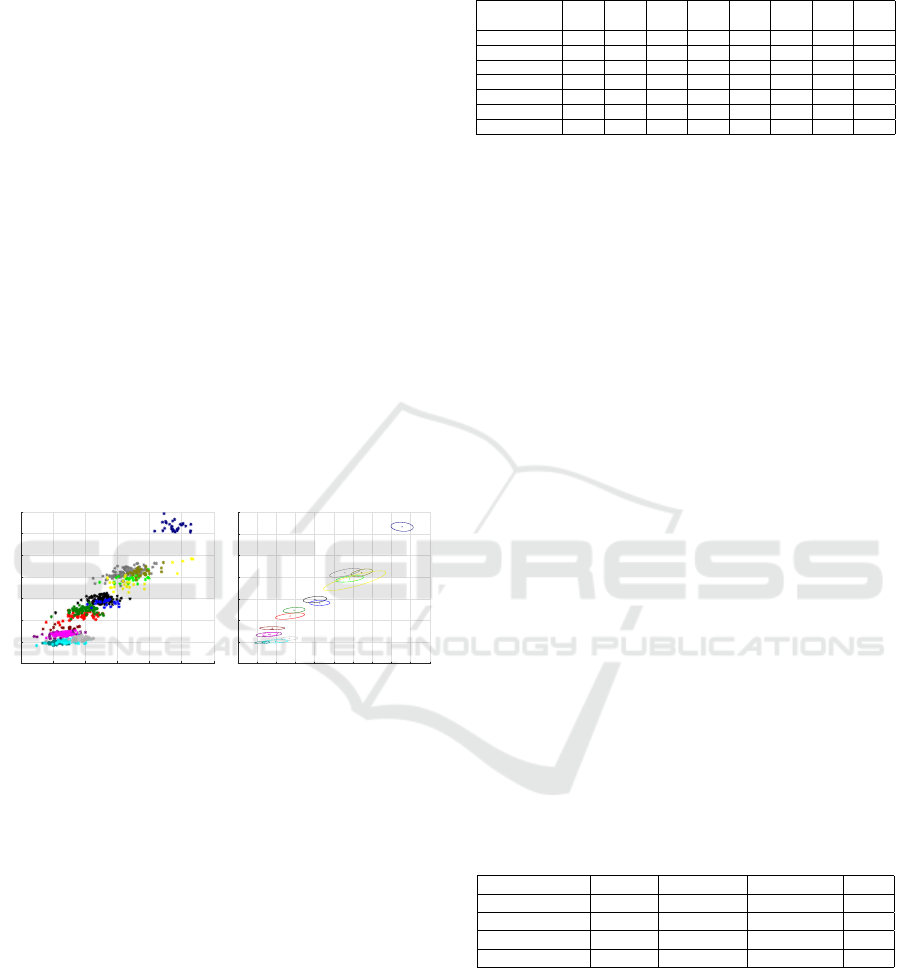
• minimum spot area s
min
= 10;
• spot dissimilarity d(p,q) = 9(x(d) − x(q))
2
+
(y(d) −y(q))
2
.
For the experiments, the original datasets collected in
the laboratory of Pushchino State Institute of Natural
Science were used. Datasets are collections of im-
ages of size 1388 ×1080 obtained from photographs
taken with a Zeiss Stemi2000 binocular microscope
equipped with an AxioCam MRc video camera. The
first set (“Day”) consists of images of entire planari-
ans, each of which was captured multiple times over
a limited period of time, not exceeding an hour. The
dataset contains a total of 1,764 images, with 28 sam-
ple classes, ranging from 16 to 113 images. The dis-
tribution of the length, understood as the length of the
main axis, and the area of the instances of the first
15 classes from the dataset are shown in Fig. 8. The
graphs show that the ability of a planarian to shrink,
stretch and bend leads to a standard deviation of up
to 10% of the mean in length, and up to 8% in area.
For this reason, even the dimensions of a planarian
are difficult to consider as a stable and discriminative
feature of the individual.
4 5 6 7 8 9 10
Length, mm
1
2
3
4
5
6
7
8
Area, mm
2
4.5 5 5.5 6 6.5 7 7.5 8 8.5 9 9.5
Length, mm
1
2
3
4
5
6
7
8
Area, mm
2
(a) (b)
Figure 8: Distribution of sizes in the “Day” dataset: (a)
individual instances and (b) standard deviations along to the
axis of PCA.
In order to investigate the influence of the num-
ber of spots—total and matched—on the identifica-
tion accuracy, a part of this dataset was taken, con-
sisting of 16 images per class. The four earliest im-
ages from the class formed a training set, the 12 most
recent were used for testing. The nearest neighbor
algorithm was utilized for identification, and the cor-
respondence of the points was established according
to formula 3, since the procedure it defines provides
the highest speed. The experimental results for a dif-
ferent number of selected spots n and matched spots
k are shown in Table 1 and allow us to conclude that
the quality of the method depends rather on the pro-
portion of the compared spots than on their absolute
number. In what follows, we set n = 24 and k = 12 as
the parameters that showed the best quality.
Table 1: Classification accuracy for the ”Day” dataset with
a different number of selected and matched spots.
P
P
P
P
P
P
P
Total
Matched
6 8 10 12 16 20 24 28
16 90.48% 95.83% 95.83% 91.96% 33.33% − − −
20 91.96% 96.43% 98.51% 98.81% 91.07% 39.29% − −
24 91.07% 95.83% 97.32% 98.81% 97.32% 88.69% 42.86% −
28 89.29% 96.13% 97.62% 98.81% 98.51% 97.32% 84.82% 45.24%
32 86.01% 91.96% 95.24% 96.73% 98.21% 98.51% 96.73% 83.93%
36 84.82% 90.18% 94.05% 96.43% 98.21% 98.21% 97.62% 95.54%
40 80.95% 87.50% 91.67% 94.35% 96.73% 98.51% 97.92% 97.32%
8.2 Comparison of Matching Methods
The full “Day” dataset was considered, 10% of the
images of each class were used as training, the rest
were used as test. The following spot matching meth-
ods discussed in the Section 6 were compared:
• assignment problem solution without additional
terms (formula 3)—AP (plain);
• assignment problem solution with L
1
-norm on the
deviation from the mean (formula 4, α = 15, β =
5)—AP (L
1
-norm);
• assignment problem solution with y-ordering pre-
served (formula 5)—AP (y-ordered);
• coherent point drift (Myronenko and Song,
2010)—CPD.
Also, the same number of SURF points was high-
lighted as points of interest. To be consistent with all
matching methods in use, only point positions were
taken, not feature vectors. The results are shown in
Table 2 and indicate the superiority of the proposed
method of spot extraction, as well as the benefit from
the use of non-trivial matching methods. Note also
that using the standard spot pattern built from train-
ing class subsample as the only class instance either
does not lead to a significant decrease in the quality
of the classification, or even slightly improves it.
Table 2: Accuracy of classification with various methods of
matching and extracting points of interest.
Method AP (plain) AP (L
1
-norm) AP (y-ordered) CPD
LoG 98.17% 98.30% 98.51% 88.97%
SURF 86.90% 87.86% 89.36% 76.19%
LoG (standards) 95.95% 96.21% 96.67% 86.55%
SURF (standards) 88.45% 89.30% 90.60% 78.00%
8.3 Observation for Several Days
To test the consistency of the proposed texture de-
scription over time, the “Week” dataset was collected,
containing photographs of 24 planaria taken on Tues-
day, Wednesday, Thursday, Friday and Monday of the
next week. At the same time, on Wednesday, Fri-
day and Monday the planaria were captured twice—
before and after noon. The dataset consists of 166
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
94
images, and the attribution of the photographs to spe-
cific individuals was established manually.
Table 3: Classification accuracy for the ”Day” dataset with
a different number of selected and matched spots.
X
X
X
X
X
X
X
X
X
X
Test day
Train day
Tu, a.m. We, a.m. We, p.m. Th, a.m. Fr, a.m. Fr, p.m. Mo, a.m. Mo, p.m.
Tu, a.m. − 68.18% 77.27% 72.72% 76.47% 58.82% 35.29% 47.06%
We, a.m. 81.82% − 95.65% 82.61% 76.47% 76.47% 52.94% 47.06%
We, p.m. 81.82% 91.30% − 95.65% 76.47% 76.47% 58.82% 52.94%
Th, a.m. 68.18% 82.61% 95.65% − 70.59% 88.24% 47.06% 29.41%
Fr, a.m. 70.59% 70.59% 76.47% 70.59% − 94.19% 58.82% 52.94%
Fr, p.m. 58.82% 70.59% 82.35% 82.35% 94.19% − 58.82% 64.71%
Mo, a.m. 47.06% 64.70% 70.59% 52.94% 64.71% 70.59% − 70.59%
Mo, p.m. 29.41% 47.06% 47.06% 35.29% 47.06% 64.71% 70.59% −
The results of the experiments are shown in Table
3 and indicate a decrease in similarity between spot
patterns over time. However, the use of time-distant
images helps to improve the quality of the classifica-
tion, which reaches the value of 92.50%, when com-
paring the test image with images taken on all other
days.
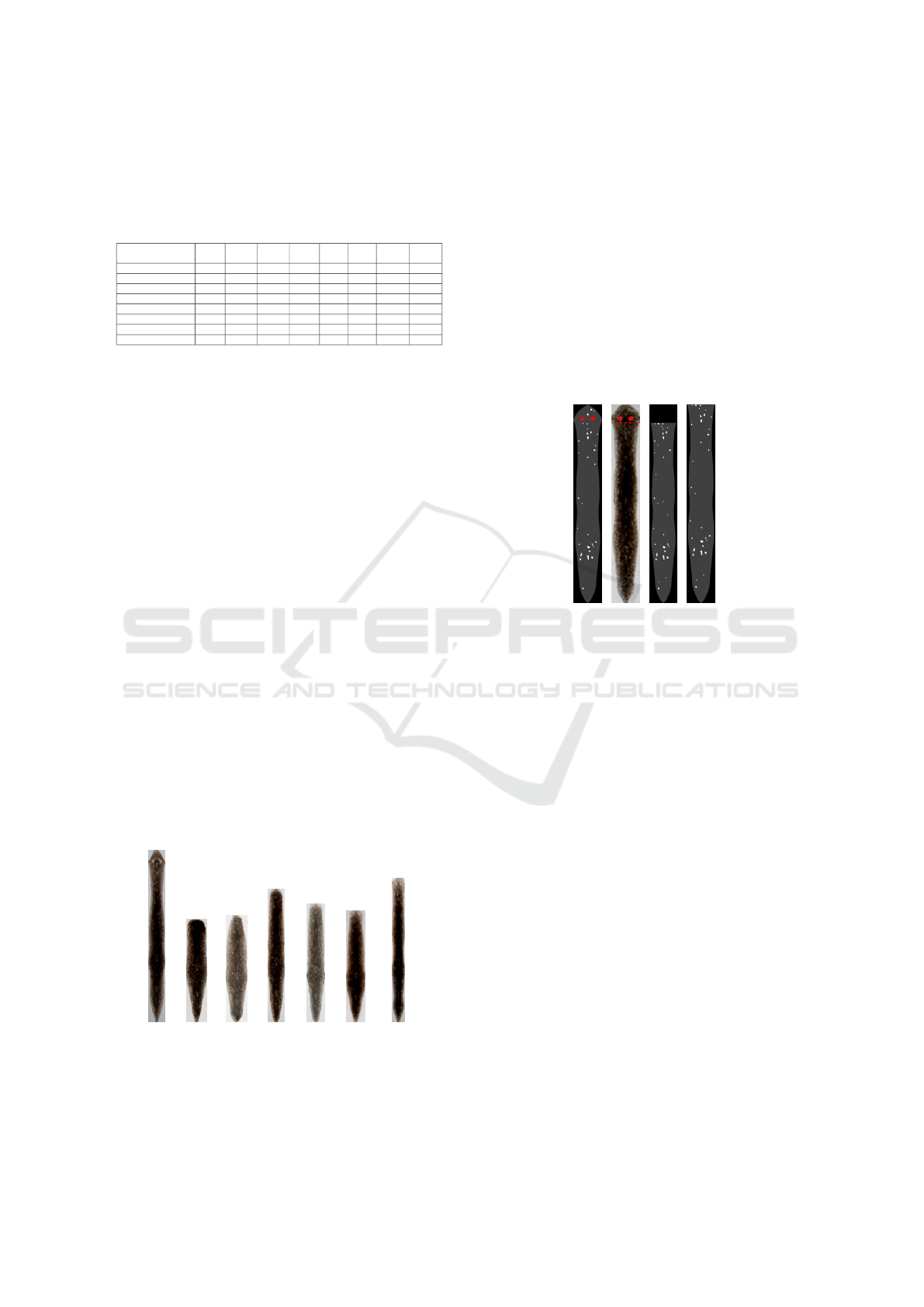
8.4 Regenerating Planaria
The “Regeneration” dataset consists of 579 images
capturing 20 planarians in the process of regeneration
after cutting off the head. Images were taken on the
first, second, third, fourth, sixth and tenth days af-
ter decapitation, and images of whole planarians be-
fore decapitation are considered as a training sam-
ple. Also, 392 images of severed heads and organisms
evolved from them were obtained, but since they have
a small length and a small number of spots, they do
not fit into the proposed model and their analysis was
left for further research. The process of regeneration
of a single planarian is shown in Fig. 9 and demon-
strates a very unpredictable shape change during the
process. It can be noted that the planarian as a whole
tends to restore its proportions, but the new spots that
have appeared in the head region do not coincide with
the old ones.
Day 0 Day 1 Day 2 Day 3 Day 4 Day 6 Day 10
Figure 9: Regeneration of planaria after cutting off the head.
To simulate the decapitation, eyes in the images
from the training sample were identified, and an as-
sumed cutting line was drawn at a level of 20 pixels
below eye level (Fig. 10ab). Further, the part of the
texture lying below this line was used for spot extrac-
tion (Fig. 10c) and the resulting pattern was stretched
in height (Fig. 10d). Such images were compared
with standards, which were considered to be the im-
ages of whole planarians before decapitation. As the
predicted cutting line may differ from the actual, the
stretching was carried out in the range from 80% to
120% of the texture height of a full planarian. The fi-
nal quality of the classification was 72.84%, the mis-
takes are explained by the instability of the planarian
morphology after decapitation and the need to predict
the dissection line.
(a) (b) (c) (d)
Figure 10: Processing the planarian after decapitation.
9 CONCLUSION
This study shows the fundamental possibility of iden-
tifying individual planarian flatworms and makes it
possible to supplement observations of groups of indi-
viduals in biological experiments by tracking individ-
ual characteristics of development and regeneration.
The problems of detecting and segmentation of an
individual planarian in the image, approximating its
shape with a fat curve based on the medial represen-
tation and extracting the texture of the planarian in the
form of a rectangle of standard size are solved. It was
shown that the texture of a planarian is described with
sufficient completeness by a set and arrangement of
light spots, which, on the one hand, are quite steadily
different between individuals, and, on the other hand,
are stable enough to persist for a rather long period of
time associated with the experiment. As directions for
further work, we can specify the processing of images
containing several animals and prediction
Identification of Planarian Individuals by Spot Patterns in Texture
95

ACKNOWLEDGEMENTS
The work was funded by Russian Foundation of Basic
Research grant No. 20-01-00664.
REFERENCES
Apyari, V., Tiras, K., Nefedova, S., and Gorbunova, M.
(2021). Non-invasive in vivo spectroscopy using a
monitor calibrator: A case of planarian feeding and di-
gestion statuses. Microchemical Journal, 166:106255.
Bagu
˜
n
`
a, J. (2012). The planarian neoblast: The rambling
history of its origin and some current black boxes.
The International Journal of Developmental Biology,
56:19–37.
Barber, B., K
¨
uhn, D., Lo, A., Osthus, D., and Taylor, A.
(2017). Clique decompositions of multipartite graphs
and completion of latin squares. Journal of Combina-
torial Theory, Series A, 151:146–201.
Bay, H., Ess, A., Tuytelaars, T., and Gool, L. V. (2008).
Speeded-up robust features (surf). Computer Vision
and Image Understanding, 110(3):346–359. Similar-
ity Matching in Computer Vision and Multimedia.
Clapham, M., Miller, E., Nguyen, M., and Darimont, C.
(2020). Automated facial recognition for wildlife
that lack unique markings: A deep learning approach
for brown bears. Ecology and Evolution, 10:12883–
12892.
Dawande, M., Keskinocak, P., Swaminathan, J. M., and
Tayur, S. (2001). On bipartite and multipartite clique
problems. Journal of Algorithms, 41(2):388–403.
Duyck, J., Finn, C., Hutcheon, A., Vera, P., Salas, J., and
Ravela, S. (2015). Sloop: A pattern retrieval engine
for individual animal identification. Pattern Recogni-
tion, 48(4):1059–1073.
Elliott, S. A. and Alvarado, A. S. (2013). The history and
enduring contributions of planarians to the study of
animal regeneration. Wiley Interdisciplinary Reviews:
Developmental Biology, 2:301–326.
Flygare, S., Campbell, M., Ross, R. M., Moore, B., and
Yandell, M. (2013). ImagePlane: An automated im-
age analysis pipeline for high-throughput screens us-
ing the planarian Schmidtea mediterranea. Journal of
Computational Biology, 20(8):583–592.
Freytag, A., Rodner, E., Simon, M., Loos, A., K
¨
uhl, H.,
and Denzler, J. (2016). Chimpanzee faces in the wild:
Log-euclidean cnns for predicting identities and at-
tributes of primates. volume 9796, pages 51–63.
Fritsch, F. N. and Carlson, R. E. (1980). Monotone piece-
wise cubic interpolation. SIAM Journal on Numerical
Analysis, 17(2):238–246.
Hughes, B. and Burghardt, T. (2017). Automated visual fin
identification of individual great white sharks. Int. J.
Comput. Vis., 122(3):542–557.
Karami, A., Tebyaniyan, H., Gooadrzi, V., and Shiri, S.
(2015). Planarians: an in vivo model for regenera-
tive medicine. International Journal of Stem Cells,
8:128–133.
Lahiri, M., Tantipathananandh, C., Warungu, R., Ruben-
stein, D. I., and Berger-Wolf, T. Y. (2011). Biometric
animal databases from field photographs: Identifica-
tion of individual zebra in the wild. ICMR ’11, New
York, NY, USA. Association for Computing Machin-
ery.
Lee, V., Ruan, N., Jin, R., and Aggarwal, C. (2010). A
Survey of Algorithms for Dense Subgraph Discovery,
pages 303–336.
Liu, C., Zhang, R., and Guo, L. (2019). Part-pose guided
amur tiger re-identification. In 2019 IEEE/CVF In-
ternational Conference on Computer Vision Workshop
(ICCVW), pages 315–322.
Lu, X., Wang, Y., Fung, S., and Qing, X. (2021). I-nema: A
biological image dataset for nematode recognition.
Mestetskiy, L. (2000). Fat curves and representation of pla-
nar figures. Computers & Graphics, 24:9–21.
Mestetskiy, L. and Semenov, A. (2008). Binary image
skeleton — continuous approach. In Proceedings
of the Third International Conference on Computer
Vision Theory and Applications - Volume 1: VIS-
APP, (VISIGRAPP 2008), pages 251–258. INSTICC,
SciTePress.
Myronenko, A. and Song, X. (2010). Point set registra-
tion: Coherent point drift. IEEE Transactions on Pat-
tern Analysis and Machine Intelligence, 32(12):2262–
2275.
Peiris, T., Hoyer, K., and Oviedo, N. (2014). Innate im-
mune system and tissue regeneration in planarians:
An area ripe for exploration. Seminars in Immunol-
ogy, 26:295–302.
Peng, H., Long, F., Liu, X., Kim, S. K., and Myers, E. W.
(2007). Straightening caenorhabditis elegans images.
Bioinformatics, 24(2):234–242.
Qu, L. and Peng, H. (2010). A principal skeleton algorithm
for standardizing confocal images of fruit fly nervous
systems. Bioinformatics, 26(8):1091–1097.
Rao, K. K., Grabow, L. C., Mu
˜
noz-P
´
erez, J. P., Alarc
´
on-
Ruales, D., and Azevedo, R. B. R. (2021). Sea turtle
facial recognition using map graphs of scales. bioRxiv.
Sheimann, I. M. and Sakharova, N. (1974). On a peculiarity
of planarian digestion. Comparative biochemistry and
physiology, 48:601–7.
Tiras, K., Mestetskiy, L., Nefedova, S., and Lomov, N.
(2021). Registration of regeneration in planarians
from photographic images. Journal of Biomedical
Photonics & Engineering.
Tiras, K., Petrova, O., Myakisheva, S., Deev, A., and
Aslanidi, K. (2015). Minimizing of morphometric
errors in planarian regeneration. Fundamental Re-
search, 2:128–133.
Tiras, K. P., Nefedova, S., Novikov, K., Emelyanenko, V.,
and Balmashov, S. (2018). In vivo methods of control
of phago- and pinocytosis on the model of planarian
digestion. In Proceedings of the International Con-
ference ”Situational Centers and Class 4i Informa-
tion and Analytical Systems for Monitoring and Secu-
rity Tasks” (SCVRT2018), volume 48, pages 346–359.
Moscow-Protvino.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
96
