
Vision Transformers for Brain Tumor Classification
Eliott Simon and Alexia Briassouli
a
Department of Data Science and Knowledge Engineering, Maastricht University, The Netherlands
Keywords:
Brain Tumor Classification, Deep Learning, Vision Transformer, Convolutional Neural Network.
Abstract:
With the increasing amount of data gathered by healthcare providers, interest has been growing in Machine
Learning, and more specifically in Deep Learning. Medical applications of machine learning range from
the prediction of medical events, to computer-aided detection, diagnosis, and classification. This paper will
investigate the application of State-of-the-Art (SoA) Deep Neural Networks in classifying brain tumors. We
distinguish between several types of brain tumors, which are typically diagnosed and classified by experts
using Magnetic Resonance Imaging (MRI). The most common benign tumors are gliomas and meningiomas,
however there exist many more which vary in size and location. Convolutional Neural Networks (CNN) are
the SoA deep learning technique for image processing tasks such as image segmentation and classification.
However, a recently developed architecture for image classification, namely Vision Transformers, have been
shown to outperform classical CNNs in efficiency, while requiring fewer computational resources. This work
introduces using only Transformer networks in brain tumor classification for the first time, and compares their
performance with CNNs. A significant difference between the two models, tested in this manner, is the lack
of translational equivariance in Transformers, which the CNNs already have. Experiments for brain tumor
classification on benchmark real-world datasets show they can achieve comparable or better performance,
despite using limited training data.
1 INTRODUCTION, RELATED
WORK
Brain tumors appear when there is an uncontrolled,
abnormal growth of cells in the central nervous sys-
tem. Although the cause of most brain tumors re-
mains unknown, experts can easily classify them in
different categories. Brain tumours are either benign
(non-cancerous), or malignant (cancerous) (Herholz,
2012). According to The Cancer Research UK, the
most common types of brain tumours are Glioma,
Meningioma, and Pituitary. Magnetic Resonance
Imaging (MRI) is a powerful non-invasive imaging
technology which allows to produce detailed anatom-
ical images of brain tumors. With the help of MRI
scans, an expert is able to determine the category of a
tumor, as well as its size and location (Scans, 2021).
With the advent of deep learning in medical imag-
ing applications, CNNs were introduced for brain tu-
mor classification in several works, achieving good
accuracy (Hossain et al., 2019), (Badza and Barjak-
tarovic, 2020). Various CNNs were examined in
(Rehman et al., 2019), in combination with very ef-
a
https://orcid.org/0000-0002-0545-3215
ficient data augmentation techniques, for brain tu-
mor classification, achieving an accuracy of 98.69%
(Rehman et al., 2019) on a 2017 dataset. However,
rapid advances in the field have led to the develop-
ment of better performing, context-aware networks,
such as Transformers, first for Natural Language Pro-
cessing (NLP), extended to computer vision.
Vision Transformer (ViT) models are usually im-
plemented for image classification or segmentation
tasks. For the sake of tumor diagnosis, ViT models
have only recently been examined, resulting in very
promising outcomes. For example, the TransBTS
(Wang et al., 2021) model allows to detect the pres-
ence of a brain tumor in a 3D MRI environment. The
model outperformed other 3D segmentation models,
reaching an accuracy of 90% (Wang et al., 2021).
Other models, such as TransMed (Dai et al., 2021),
which consists of a combination of Transformer and
CNN, have further improved the quality of tumor di-
agnosis. The reason for combining the two archi-
tectures is that most tumor classification datasets are
small, while the efficiency of transformer networks
still highly depends on the amount of data used for
training (Dai et al., 2021).
Simon, E. and Briassouli, A.
Vision Transformers for Brain Tumor Classification.
DOI: 10.5220/0010834300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 123-130
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
123

In this paper, we examine for the first time the de-
tection and classification of tumors from a recent MRI
Brain tumor dataset (MRI Kaggle dataset, 2020) us-
ing ViTs alone, that are trained from scratch. The
SoA on this dataset is based on CNNs, and attained an
accuracy of 95.40% (Badza and Barjaktarovic, 2020),
while ViT’s have not been applied to it. Specifically,
at the time of writing this paper, VIT’s alone had not
been applied to the problem of brain tumor detection
and classification on related datasets, including the re-
cent only used here (MRI Kaggle dataset, 2020).
Unlike past works, our approach relies solely on
transformers, trained from scratch on this relatively
small dataset. We compare their performance to that
of a CNN, which we designed and trained for this
dataset, demonstrating that they outperform it as well.
Overall, ViT’s are demonstrated to perform very well,
despite lacking in inductive biases, translational in-
variance and equivariance that characterize CNNs,
while being trained from scratch on a relatively small
training dataset. This indicates that ViTs alone can be
used for tumor detection and classification, but also -
indirectly - shows the role of spatial attention in ViT
vs. translational invariance that is present in CNNs.
Data augmentation was also examined, but shown to
require more computational resources, in order for
the augmented datasets to be appropriately leveraged,
as in (Rehman et al., 2019), leading us to conclude
its potential for improving results is possible when
higher computational resources are available.
This paper is structured as follows: Section 2 de-
scribes the dataset that is used for classification, and a
binarized version we create for detection. The meth-
ods used are presented in Section 3, where a descrip-
tion of the general Transformer Network architecture
is given, followed by its application in Computer vi-
sion, known as Vision Transformers, and the CNN
used in this work for comparison. Experimental re-
sults are presented in Sections 4 and section 5 presents
our conclusions and future research directions.
2 DATASET
We consider a recent (2020) data set of Magnetic Res-
onance Imaging (MRI) annotated data, where each
image depicts either a type of brain tumor (one of
three types), or no tumor (MRI Kaggle dataset, 2020).
Some characteristic images from it are shown in
Fig. 1. Detecting and classifying these tumors is a
typical classification problem, therefore a lot of data
is required in order to build a robust model (He et al.,
2016). A rule of thumb is that 1000 of images per
class is already enough (Warden, 2017).
Figure 1: Sample images from the benchmarking data set.
Figure 2: Number of images per each category.
Our testing dataset contains 100 images of glioma
tumors, 115 of meningioma tumors, 74 of pituitary
tumors and 105 with no tumor. Our training dataset
contains 826 images of glioma tumors, 822 of menin-
gioma tumors, 827 of pituitary tumors and 395 with
no tumor. The dataset does not contain any external
information about the patients, therefore its applica-
tion is restricted to image classification. The distri-
bution of the data among the different classes is quite
balanced, with a lower amount of no tumor images
available for training, as shown in Fig. 2.
2.1 Binary Data Set
In order to examine the performance of ViT’s on the
detection problem, we first consider the simpler prob-
lem of detecting the presence of a tumor versus no
tumor. To this end, we created a binary data set
from (MRI Kaggle dataset, 2020), which contains
BIOIMAGING 2022 - 9th International Conference on Bioimaging
124
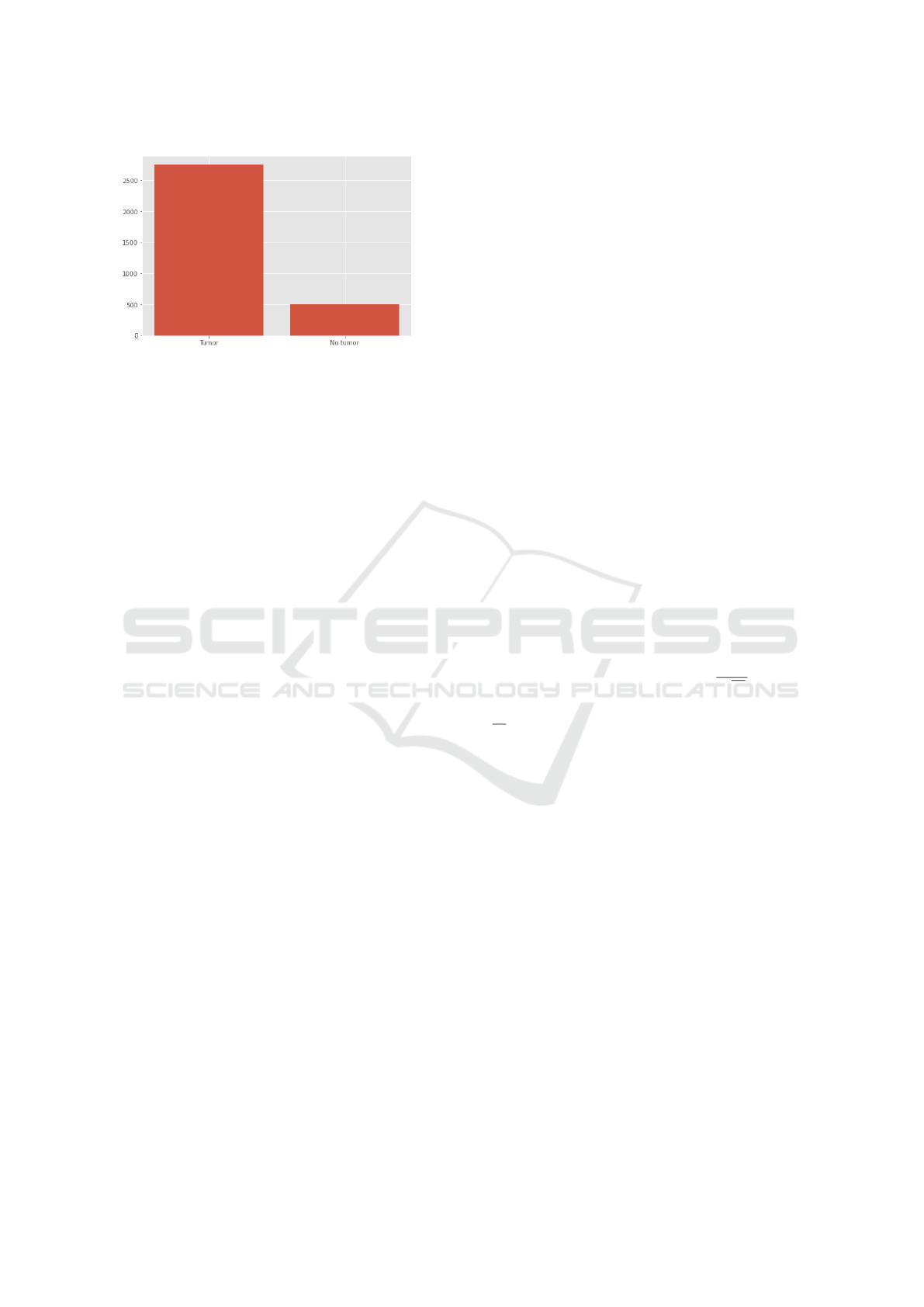
Figure 3: Distribution of Binary Dataset.
only two classes. The idea is to merge the glioma,
meningioma, and pituitary into a single tumor-class.
The other class contains the images of a brain with
no brain tumor. The resulting binary dataset is quite
unbalanced, because the images with tumors have far
more data available for training (Fig. 3), however it is
expected to suffice for the binary detection problem.
For both the binary and original data split, data
augmentation is a plausible approach to addressing
dataset imbalance and improving performance. For
our kind of data, data augmentation needs to be ap-
plied carefully so as to not modify informative pixel
values in the images, so we only tested different
kinds of image rotation. Our experiments showed
that, for this data, rotation did not improve results,
indicating that it did provide sufficient variation in
the augmented training data, so we did not pursue it
further. Effective data augmentation would require
higher computational resources and extensive train-
ing, in order to sufficiently leverage the additional in-
formation in the augmented data and explore its effect
in depth, which is beyond the scope of this work.
3 METHODS
3.1 Transformer Network
Transformer Networks became the SoA technique for
many Natural Language Processing tasks, such as ma-
chine translation, or text summarization (Wang et al.,
2019). Similarly to Recurrent Neural Networks, the
input data must be sequential. However, the novel
transformer model uses parallelization, which consid-
erably reduces the training time.
The core of the model consists of an encoder-
decoder architecture (Vaswani et al., 2017). In Recur-
rent Neural Networks, the encoder generates an em-
bedding for each word, one at a time. However, each
instance of a word depends on the previously embed-
ded words, which leads to very inefficient results for
large texts (Cho et al., 2014). In transformer mod-
els this issue is surpassed, as the encoder of a trans-
former model captures the entire sequence and gen-
erates an embedding for each word simultaneously.
Each of these embeddings consists in a vector that
encapsulates the meaning of the word. Therefore,
similar words have closer numbers in their vectors
(Zichao and et al., 2016). Since similar words may
have different meanings, the model uses a positional
encoder, which provides some context, based on the
positions of the words in the sentence. Thereafter,
the embedding vectors that contain context about the
words are fed into the encoder block. The first step
of this encoder block involves attention, which deter-
mines the most important parts of the input (Vaswani
et al., 2017), (Cho et al., 2014). An attention vector
is assigned to every word, which captures the contex-
tual relationship between the given word and the other
words in the sentence (Koner et al., 2020). Then, each
attention vector is fed into a feed forward network,
such that it is reusable for the next encoding, or de-
coding block. After each sub-layer, the input is nor-
malized and reduced to an exactly one dimensional
vector for each word. The decoder has the same ini-
tial steps, however, the self attention sub-layer uses a
masking operation. The attention is computed as fol-
lows: (Vaswani et al., 2017):
Attention(Q, K, V ) = so f tmax(
QK
T
√
d
k
)V, (1)
where Q are Queries, K are keys, V are values and
√
d
k
is the square root of the dimension of the keys.
QK
T
allows to compute the similarity between the
words in Q and K. In the decoder, queries come
from the target words, and both keys and values come
from the original words. Since the transformer works
with word embeddings, there is no time dependency.
Henceforth, we must perform a pointwise operation
between QK
T
and a masked matrix, in such a way that
the words are blocked from attending future words
during the training (Vaswani et al., 2017). After-
wards, the encoder-decoder attention layer generates
similar attention vectors for words in both the input
and the output vocabulary. The linear sub-layer is an-
other feed forward neural network which is used to
expand the dimension into the number of words in
the target vocabulary. The softmax function maps the
input to a probability distribution, which is human-
interpretable. The output of the decoder is the word
with the highest probability.
Vision Transformers for Brain Tumor Classification
125
3.2 Vision Transformers
Convolutional Neural Networks (CNNs) are very effi-
cient models for computer vision tasks, and still make
up the SoA for tumor classification on the benchmark
dataset examined in this work (MRI Kaggle dataset,
2020). Recently, researchers have been attempting to
improve their performance by combining them with
the self-attention architectures (Yamashita, 2017). Vi-
sion transformers, which incorporate attention, were
introduced in 2020, and presented two main achieve-
ments (Dosovitskiy et al., 2021). First of all, the train-
ing time of the model is 80% faster than the Noisy
Student for the same accuracy, according to the Im-
ageNet benchmark (ImageNet, 2007). Secondly, the
model does not rely on convolutions, but only on self-
attention. For computer vision, the attention needs
to be evaluated between pixels. However, computing
the relationship between the pixels of a 520x520 im-
age would require 270,000 combinations, so comput-
ing the attention for each of the combinations would
be computationally very expensive. Besides, in most
cases, a pixel on the bottom left corner of an im-
age does not have a strong relationship with the pixel
on the top right corner. Vision Transformers over-
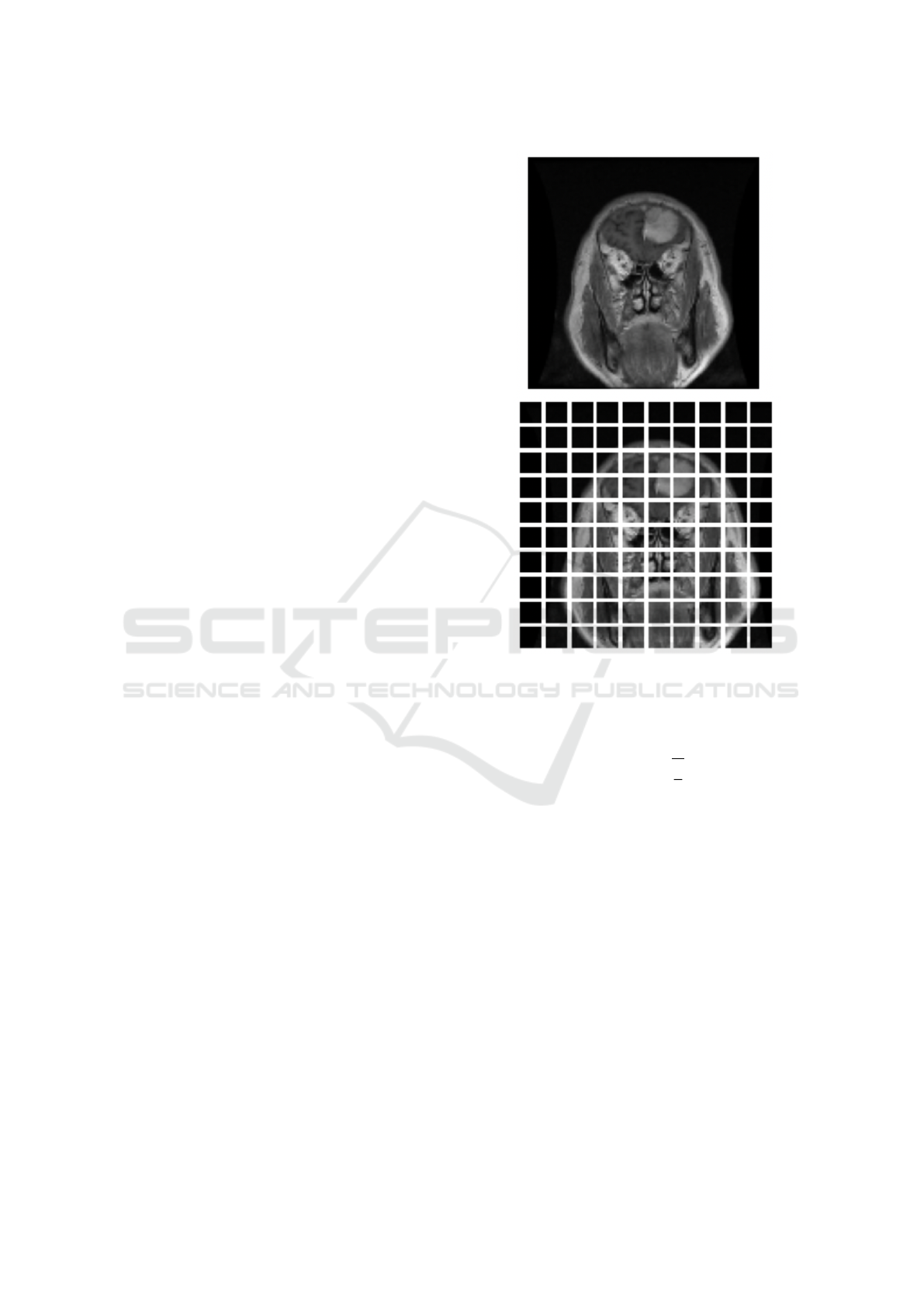
come this problem by splitting the images into sev-
eral equal-sized patches (Dosovitskiy et al., 2021),
thus examining more spatially relevant and informa-
tive pixels instead of the entire image.
Each patch is simultaneously embedded, and a po-
sitional embedding is also applied to each of them.
This positional embedding injects important infor-
mation about the absolute or relative position of the
image patches in the “sequence” (image), in Eq(2).
Thereafter, the embedded patches are fed into the
transformer encoder block. This encoder block con-
sists of a Multi-Head self-attention sub-layer which
follows a normalization layer. A skip-connection
layer is added, in order to allow gradients to flow di-
rectly through the network (He et al., 2016). Finally, a
Multi-Layer-perceptron (MLP) allows for classifica-
tion. The MLP is surrounded by both a normalization
and a skip-connection layer.
z
0
= [x
class
;x
1
p
E; x
2
p
E; ...x
N
p
E] + E
pos
, E ∈ R
(P
2
.C)∗D
(2)
z
0
l
= MSA(LN(z
l−1
)) + z
l−1
, l = 1....L (3)
z
l
= MSP(LN(z
0
l
)) + z
0
l
, l = 1....L (4)
y = LN(z
0
L
) (5)
where MSA stands for Muli-Head Self Attention, and
LN is the Layer Norm (Dosovitskiy et al., 2021).
The MLP layer uses the Gaussian Error Linear Unit
Figure 4: Splitting the image into patches.
(GELU) activation function. The GELU function is
computed as follows:
GELU (x) = 0.5x(1 + tanh(
r
2
π
(x + 0.044715x
3
)))
(6)
The main advantage of GELU is that it avoids van-
ishing gradients problem (Hendrycks and Gimpel,
2016). Most recent transformer network models, such
as BERT, or GPT-2 use this activation function (De-
vlin et al., 2019), (Radford and et al., 2019).
A block diagram showing the patch-based Vision
Transformer used in this work for tumor detectino and
classification is shown in Fig. 5.
3.3 CNN Model
Most current image classification tasks for medical
applications that involve deep learning rely on Convo-
lutional Neural Networks (Yamashita, 2017), (Koner
et al., 2020), (Dosovitskiy et al., 2021), with newer
ones only recently combining Transformers with
BIOIMAGING 2022 - 9th International Conference on Bioimaging
126
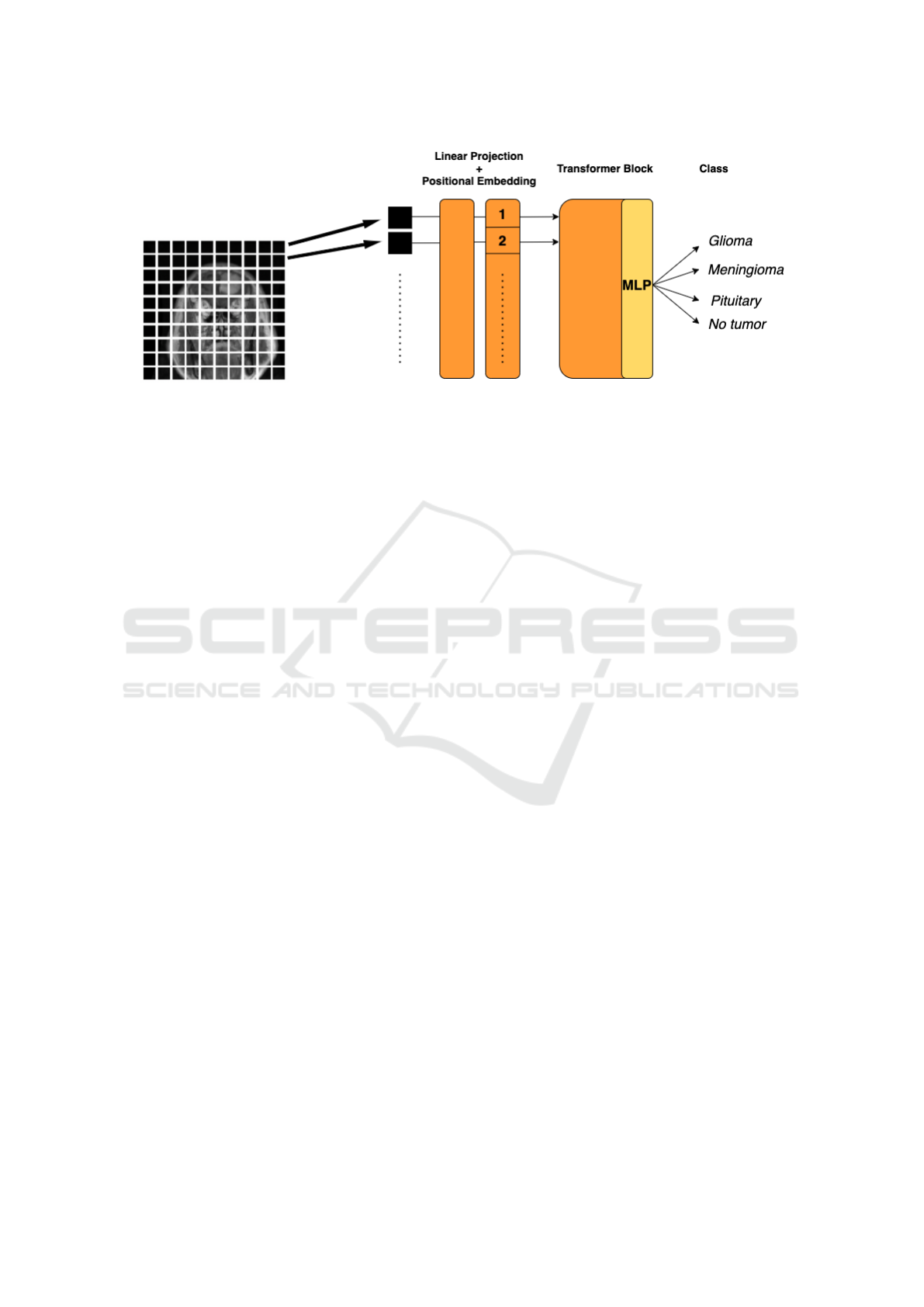
Figure 5: Vision Transformer (ViT) Architecture for detection/classification of MRI brain tumors.
CNNs (Wang et al., 2021). In order to test the effi-
ciency of ViT-alone compared to CNN-alone, we con-
struct a CNN network for our dataset and compare its
performance to that of ViT. Various CNN architec-
tures were tested, and after appropriate hyperparame-
ter optimization and experimentation we used the one
in Fig. 6 that led to the best results.
We compare our ViT results with those of our
CNN, and indirectly compare with the SoA CNN for
brain tumor classification (Badza and Barjaktarovic,
2020). The CNN of (Badza and Barjaktarovic, 2020)
is not directly comparable with ours, as their CNN
involves more convolutional layers, the dataset split
they use is not known, and also carry out data aug-
mentation and k-fold validation. In our case, we only
implement our CNN to compare its “barebones” per-
formance with that of the ViT under the same condi-
tions (same dataset, same train/test split, no augmen-
tation). In this manner, we aim to objectively and
clearly showcase the effect of context and attention,
under the same experimental conditions.
4 EXPERIMENTS
In order to compare the efficiency of ViT-alone com-
pared to CNN-alone, we perform experiments for the
same train-test split on the data set, as explained
above. The idea is to train both models on 80% of
the data, while keeping the rest for validation/testing
purposes, so we used an 80/20 training/validation
split (Russell and Norvig, 2009). The accuracy of
the model is the percentage of correctly classified in-
stances. In order to compute it, we need to divide the
sum of the True Positive and True Negative terms by
the total number of testing instances. Another inter-
esting measure, for both binary and multi-class prob-
lem, is the confusion matrix. Indeed, the confusion
matrix shows, among all the possible classes, what
the predicted value is. The diagonal elements from
the matrix represent instances that have been correctly
classified.
4.1 Validation/Training
As a first step in examining the performance of our
two models, we carried out validation experiments.
The validation set allows us to observe that the ViT
model is overfitting the data to a small degree, but
training loss remains very close to the validation loss,
or lower, making this a minimal effect. The CNN
slightly overfits the dataset (Fig. 8), which can be
attributed to the limited training data. Figs. 9, 10 with
the validation/training accuracies shows this is still
the case, but is not a significant effect.
4.2 Classification Performance
We examine the performance of both architectures
for the problem of tumor classification for the four
classes of tumors Glioma, Meningioma, No Tumor,
Pituitary. The confusion matrix in Table 1 shows that
the ViT indeed accurately finds the classes with low
false positives. Table 2 shows it results in overall
accuracy of 96.5 %, and surpasses the CNN, which
achieves an accuracy of 89.78 %.
The current SoA on the same data achieves a 95.4
% classification accuracy (Badza and Barjaktarovic,
2020) using a CNN-based approach. It should be
noted that we only indirectly compare our results to
theirs, as they do not make their code and all imple-
mentation details available. They also perform data
augmentation, increasing their CNN-based accuracy
to 96.4 %, which is very close to the accuracy we
achieved using ViTs. However, in the case of specific
applications like medical imaging, data augmentation
needs to be carefully applied, so as to not distort cru-
cial information in the medical images.
Vision Transformers for Brain Tumor Classification
127

Figure 6: CNN model compared with Vision Transformers.
We carried out initial testing with data augmenta-
tion and observed that it can also worsen accuracy by
introducing unexpected distortions, and requires sig-
nificantly more training time to achieve a decent ac-
curacy. This demonstrates that augmentation needs
to be implemented strategically to avoid such issues,
while it also entails a much higher computational
Figure 7: Model loss over time for the ViT for the validation
and training data.
Figure 8: Model loss over time for the CNN for the valida-
tion and training data.
cost, which increases even more when k-fold valida-
tion is involved. For these reasons, and in order to
avoid a computationally costly solution, we do not
proceed with data augmentation in these experiments,
and show we still achieve very high accuracies that
surpass our CNN under the exact same setup. Some
examples of correctly classified tumors by the ViT can
be seen in Fig. 11.
Table 1: Vision Transformer Confusion matrix for Classes:
1: Glioma, 2: Meningioma, 3: No Tumor, 4: Pituitary.
Class 1 2 3 4
1 139 5 3 0
2 2 172 1 0
3 1 0 76 0
4 0 0 0 178
4.3 Detection Performance (Binary
Case)
We also examine the performance of the CNN and
the ViT for the binary dataset containing samples of
BIOIMAGING 2022 - 9th International Conference on Bioimaging
128
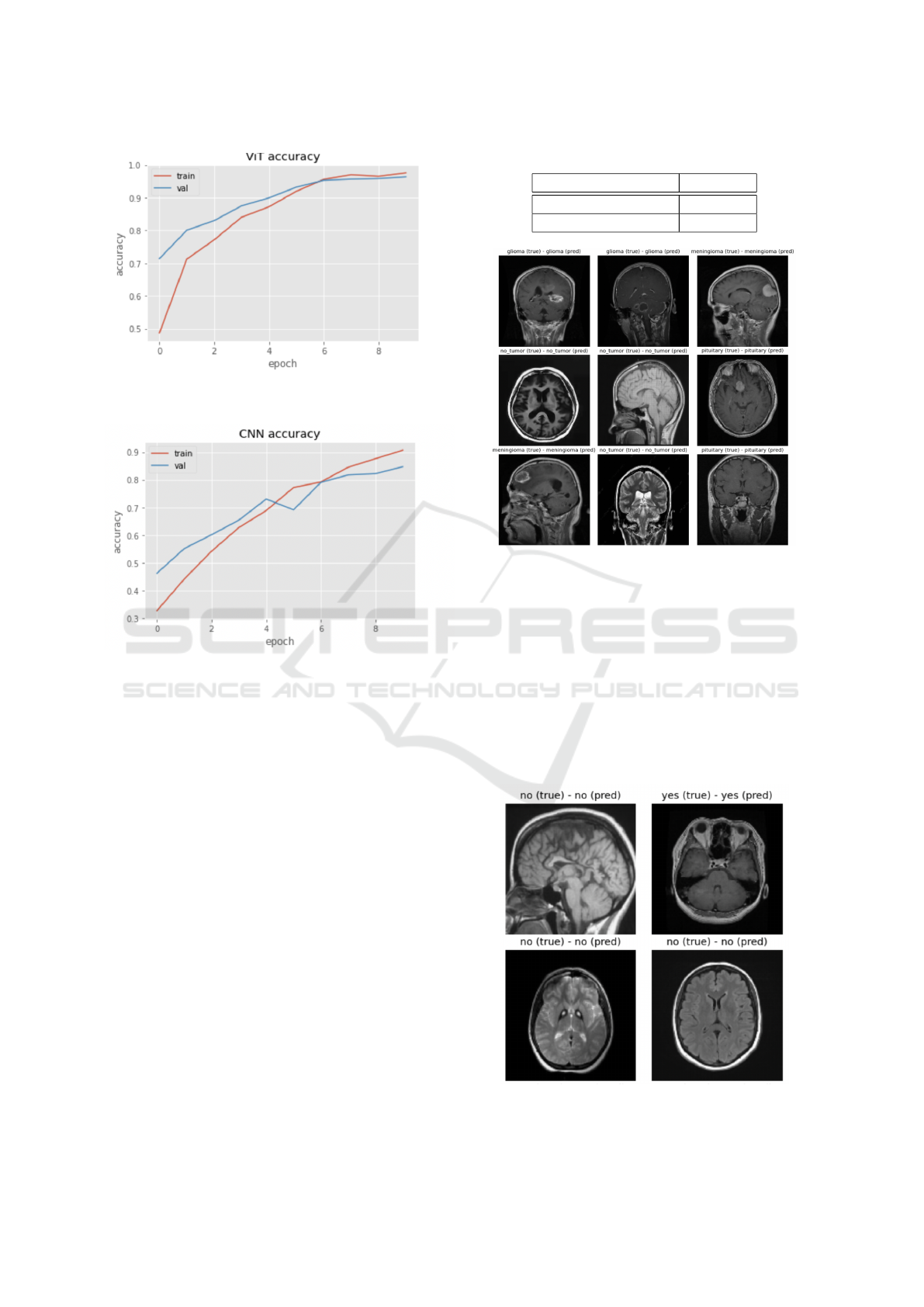
Figure 9: Model accuracy over time for the ViT for the val-
idation and training data.
Figure 10: Model accuracy over time for the CNN for the
validation and training data.
tumor vs no tumor. In this case, the confusion matrix
of Table 3 again shows the ViT correctly detects most
tumor/no tumor cases, with a few false alarms that
show it still does not overfit. Fig. 12 shows character-
istic samples that are correctly labeled as tumor/no tu-
mor by the ViT. In this simpler task, the ViT performs
very well, achieving an exceptionally high accuracy
of over 98 %, despite the dataset being unbalanced.
Although the binary classification task can be con-
sidered simpler than that of four-class classification
examined above, its large imbalance could introduce
errors to our data, such as missing the no tumor cases.
These good results can be attributed to the fact that at-
tention in ViT’s helps them focus on salient regions of
each image, achieving higher detection accuracy and
fewer false alarms introduced from other regions.
5 CONCLUSIONS
This research proposed applying recently introduced
Vision Transformer models to the challenging prob-
lems of brain tumor detection and classification on a
Table 2: Final Accuracies for tumor classification.
Classification Model Accuracy
ViT 0.965
CNN 0.8978
Figure 11: ViT Model Prediction/Actual Label for tumor
classification.
benchmark dataset. Vision Transformers were tested
as-is, i.e. without any convolutional layers, so as
to examine the effect of their spatial attention alone,
without the aid of translational invariance present in
CNNs. We trained the ViT from scratch on a bench-
marking dataset of relatively small size, that is quite
unbalanced, and avoided adding data augmentation
or cross-validation to examine its performance as-is,
and to reduce computational requirements. The ViT
model performed extremely well, also compared to
Figure 12: Binary case: ViT Model Prediction/Actual La-
bel.
Vision Transformers for Brain Tumor Classification
129

Table 3: Vision Transformer Confusion matrix for the Bi-
nary Model (Tumor/No tumor).
Tumor Detection no tumor tumor
no tumor 99 7
tumor 2 497
our custom-built CNN trained under the same exact
conditions. Although the model did not train on a
huge amount of data, and used an unbalanced it still
managed to achieve 96.5 % classification accuracy,
and over 98 % detection accuracy, which is impres-
sive. We compared to CNNs, which are used in the
SoA for such tasks, and demonstrated that the ViT
can still achieve better accuracy, despite lacking trans-
lational invariance. Some modifications could im-
prove the efficiency of the model, such as optimizing
the hyper-parameters. Adding another regularization
technique and appropriate data augmentation could
also ensure the model does not overfit the data. These
solutions entail another tradeoff, as they are likely to
significantly increase training time to achieve good
accuracy. Finally, future work includes investigating
the use of recently introduced Convolutional Vision
Transformers (CvT), which attain higher results than
normal Vision Transformers.
REFERENCES
Badza, M. and Barjaktarovic, C. (2020). Classification of
brain tumors from mri images using a convolutional
neural network. Applied Sciences.
Cho, K., van Merrienboer, B., G
¨
ulc¸ehre, C¸ ., Bahdanau, D.,
Bougares, F., Schwenk, H., and Bengio, Y. (2014).
Learning phrase representations using RNN encoder-
decoder for statistical machine translation. In Mos-
chitti, A., Pang, B., and Daelemans, W., editors, Pro-
ceedings of the 2014 Conference on Empirical Meth-
ods in Natural Language Processing, EMNLP 2014,
October 25-29, 2014, Doha, Qatar, A meeting of
SIGDAT, a Special Interest Group of the ACL, pages
1724–1734. ACL.
Dai, Y., Y. Gao, Y., and Liu, F. (2021). Transmed: Trans-
formers advance multi-modal medical image classifi-
cation. Diagnostics.
Devlin, J., Chang, M., Lee, K., and Toutanova, K. (2019).
BERT: pre-training of deep bidirectional transformers
for language understanding. In Burstein, J., Doran,
C., and Solorio, T., editors, Proceedings of the 2019
Conference of the North American Chapter of the As-
sociation for Computational Linguistics: Human Lan-
guage Technologies, NAACL-HLT 2019, Minneapolis,
MN, USA, June 2-7, 2019, Volume 1 (Long and Short
Papers), pages 4171–4186. Association for Computa-
tional Linguistics.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., Uszkoreit, J., and Houlsby,
N. (2021). An image is worth 16x16 words: Trans-
formers for image recognition at scale. In Interna-
tional Conference on Learning Representations.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Hendrycks, D. and Gimpel, K. (2016). Gaussian error linear
units (gelus). arXiv preprint arXiv:1606.08415.
Herholz, K. (2012). Brain tumors. PubMed.
Hossain, T., Tonmoy, F., Shishir, S., Ashraf, M., Nasim, A.,
and Shah, F. (2019). Brain tumor detection using con-
volutional neural network. In 2019 1st International
Conference on Advances in Science, Engineering and
Robotics Technology (ICASERT), pages 1–6.
ImageNet (2007). Imagenet benchmark (image classifica-
tion), 2007. https://paperswithcode.com/sota/image-
classification-on-imagenet.
Koner, R., Sinhamahapatra, P., and Tresp, V. (2020). Rela-
tion transformer network. CoRR, abs/2004.06193.
MRI Kaggle dataset (2020). Brain tu-
mor classification (mri), 2020.
https://www.kaggle.com/sartajbhuvaji/brain-tumor-
classification-mri.
Radford and et al. (2019). Language models are unsuper-
vised multitask learners. OpenAI.
Rehman, A., Naz, S., Razzak, I. M., Akram, F., and Im-
ran
˚
a, M. (2019). A deep learning-based framework
for automatic brain tumors classification using trans-
fer learning. Circuits, Systems, and Signal Processing.
Russell, S. J. and Norvig, P. (2009). Artificial Intelligence:
a modern approach. Pearson, 3 edition.
Scans (2021). Best scans to detect cancer.
https://www.envrad.com/best-scans-to-detect-cancer/.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A., Kaiser, L., and Polosukhin, I. (2017).
Soft-gated warping-gan for pose-guided person image
synthesis. Advances in Neural Information Processing
Systems, pages 5998–6008.
Wang, Q., Li, B., Xiao, T., Zhu, J., Li, C., Wong, D., and
Chao, L. (2019). Learning deep transformer models
for machine translation. In 57th Annual Meeting of
the Association for Computational Linguistics (ACL)
2019, pages 1810–1822.
Wang, W., Chen, C., Ding, M., Li, J., Yu, H., and Zha, S.
(2021). Transbts: Multimodal brain tumor segmen-
tation using transformer. In 24th International Con-
ference on Medical Image Computing and Computer
Assisted Intervention (MICCAI 2021).
Warden (2017). How many images do
you need to train a neural network?
https://petewarden.com/2017/12/14/how-many-
images-do-you-need-to-train-a-neural-network/.
Yamashita, R. (2017). Convolutional neural networks: an
overview and application in radiology. Insights into
Imaging.
Zichao, Y. and et al. (2016). Hierarchical attention net-
works for document classification. In Proceedings of
the 2016 Conference of the North American Chapter
of the Association for Computational Linguistics: Hu-
man Language Technologies, page 1480–1489.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
130
