
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A
Multilinear Approach
Metin Calis
1 a
, Massimo Mischi
2 b
, Alle-Jan Van Der Veen
1 c
and Borb
´
ala Hunyadi
1 d
1
Faculty of Electrical Engineering, Mathematics and Computer Science, Delft University of Technology,
2628 CD Delft, The Netherlands
2
Department of Electrical Engineering, Eindhoven University of Technology, 5600 MB Eindhoven, The Netherlands
Keywords:
Multilinear Singular Value Decomposition, Dynamic Contrast-enhanced Ultrasound, Prostate Cancer, Tensor
Decomposition.
Abstract:
The recent advances in three-dimensional imaging of contrast-enhanced ultrasound acquisitions enable the
characterization of the tissue with a single intravenous injection of microbubbles. Many cancer markers have
been extended to cover for the three-dimensional contrast ultrasound. However, most of the signal denoising
algorithms do not exploit the added dimensionality and vectorize the spatial dimensions, causing a loss of
information about the location of the voxels. This paper proposes a denoising algorithm based on the mul-
tilinear singular value decomposition and compares it to the singular value decomposition. The ranks are
estimated based on information-theoretic criteria, and improved performance has been observed for modeling
the time-intensity curves.
1 INTRODUCTION
Prostate cancer (PCa) is found to be the most com-
mon malignancy among American men except for
skin cancer, and according to the estimates for 2021,
248, 530 new cases are expected of which 34, 130
people will die from the disease (Siegel et al., 2021).
The recommended guidelines for detection and grad-
ing of PCa is ≥ 10-core systematic biopsy (Mottet
et al., 2017), which consists of an invasive procedure
that can still miss significant PCa lesions (Ukimura
et al., 2013). In addition, infectious complications
might arise due to the insertion of needles through
the rectal wall (Loeb et al., 2013). Also, because
of the poor patient selection by prostate specific anti-
gen blood testing, about 3 of 4 biopsies are in retro-
spect unnecessary, as no cancer is found. Recent ad-
vances in imaging techniques can potentially reduce
the needle-cores in systematic biopsy and overcome
unnecessary biopsies (Elwenspoek et al., 2019).
One of the featured techniques to classify PCa
is the detection of angiogenesis, which is the rapid
a
https://orcid.org/0000-0002-7576-6647
b
https://orcid.org/0000-0002-1179-5385
c
https://orcid.org/0000-0003-4249-585X
d
https://orcid.org/0000-0002-9333-9024
growth of new blood vessels around the tumourous
region. The newly formed capillaries create a tor-
tous and a highly irregular network, that can be an-
alyzed by modelling the flow dynamics of the ul-
trasound contrast agents (UCAs) (Seitz et al., 2009)
(Russo et al., 2012). Although several modalities have
been proposed that can monitor the UCAs, we focus
on dynamic contrast-enhanced ultrasound (DCEUS)
because of the promising results for PCa localization
(Liu et al., 2020) (Wildeboer et al., 2019). In DCEUS,
contrast agents that are smaller than 10 µm are imaged
using a low energy ultrasound pulse. The frequency
of the pulse is around the resonance frequency of
the UCAs, which enhances the contrast between the
microbubbles and the surrounding tissue. The time-
intensity curves (TICs) of these contrast agents corre-
lates with the underlying vasculature (Strouthos et al.,
2010), which can be exploited to classify PCa.
There has been a substantial amount of work in
the analysis of the perfusion characteristics of TICs
(Eckersley et al., 2002) (Elie et al., 2007). Al-
though increased perfusion is expected due to the in-
creased microvascular density, contradictory effects
of angiogenesis and perfusion have been observed
(Gillies et al., 1999). The flow resistance might re-
duce due to the introduction of arteriovenous shunts
and lack of vasomotor control. This effect might
192
Calis, M., Mischi, M., Van Der Veen, A. and Hunyadi, B.
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A Multilinear Approach.
DOI: 10.5220/0010838000003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 4: BIOSIGNALS, pages 192-199
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

not be observed due to the small diameter of the
newly formed microvessels and the increased intersti-
tial pressure because of the extravascular leakage (De-
lorme and Knopp, 1998). A different path has been
taken by (Mischi et al., 2009) (Mischi et al., 2003)
where the authors have modeled the multi-trajectory
bubble transport inside the prostate as a convective-
dispersion process and introduced a new model based
on dispersion characteristics, namely the modified lo-
cal density random walk model (mLDRW) (Kuenen
et al., 2011). The authors have reported a good cor-
relation between dispersion and angiogenesis even
though the single-dimensional TICs suffer from a low
signal-to-noise (SNR) ratio.
Several techniques were proposed to remove the
noise of various origins from the DCEUS signals.
Spatial filters were used to denoise the speckle noise
(Bar-Zion et al., 2015) (Joel and Sivakumar, 2018),
temporal filters were proposed to denoise the clut-
ter (Bjaerum et al., 2002). These methods assumed
that the temporal and spatial frequency of the de-
sired signal and the noise were different. Using a
different approach, the authors at (Wildeboer et al.,
2020) proposed the blind source separation (BSS)
of the DCEUS recording into sources where the de-
sired signal was recovered by discarding the noise
sources. The best performing BSS was found to be
singular value decomposition (SVD) for modelling
of the time intensity curves. The dynamics of the
UCAs have been captured in the first few singular val-
ues of the decomposition. Although several cancer
markers such as the similarity between TICs (Schalk
et al., 2015a) and solutions to convective dispersion
models (Wildeboer et al., 2018b) (Wildeboer et al.,
2018a) have been extended to 3D, the denoising algo-
rithms do not take the multidimensional structure of
the recording into consideration.
The SVD denoising analyzed by (Wildeboer et al.,
2020) creates the Casorati matrix, which vectorizes
the spatial dimension into rows and the temporal di-
mension into columns. The vectorization of the spa-
tial dimension into rows results in a loss of spatial
information regarding the location of the voxels. The
multilinear singular value decomposition (MLSVD)
proposed by (De Lathauwer et al., 2000) keeps the
tensor format of the data and generalizes the concept
of SVD to multiple dimensions. The information that
is retained by keeping the tensor format of the data is
hypothesized to enable a better representation of the
bubble dynamics and hence, a better denoising capa-
bility. The MLSVD has been applied for clutter filter
denoising in power Doppler images (Zhu et al., 2020)
(Ozgun and Byram, 2020). Improved sensitivity and
specificity have been observed. As far as the authors’
knowledge, there has not been any work done in ana-
lyzing the performance of MLSVD in TIC dispersion
modeling. This paper aims to answer the following
research question: Can the retained 3D structural in-
formation provided by MLSVD improve the classifi-
cation performance of TIC dispersion modeling com-
pared to SVD?
2 BACKGROUND INFORMATION
In this section, the notation is introduced and the mul-
tilinear singular value decomposition (MLSVD) is ex-
plained.
2.1 Notation
The tensor notation of (De Lathauwer et al., 2000) is
adapted in this paper. Tensors are represented with
caligraphic letters, for example, Y and G. The matri-
ces are represented with bold face letters, for example,
U
(1)
and V. Here, the numbers given as superscripts
in parenthesis are used to refer to the different matri-
ces, that share a similar property. An example could
be the n−mode factor matrices of the same decompo-
sition, which will be explained in the following sub-
section. The scalars are represented with lower case
letters, such as, (A)
i j
= a
i j
and (U)
i
1
i
2
i
3
= u
i
1
i
2
i
3
. The
subscripts refer to the indices in different modalities.
For example, a
i j
refer to the ith row and the jth col-
umn of the matrix A.
2.2 MLSVD
Definition 1: The casorati matrix is generalized by
the n − mode unfoldings of a tensor. The 1 − mode
unfolding of tensor
Y ∈ R
N
x
×N
y
×N
z
×N
t
is represented as Y
(1)
∈
R
N
x
×N
y
N
z
N
t
where the columns are the infor-
mation in x direction. Likewise, the 2 − mode
unfolding is Y
(2)
∈ R
N
y
×N
x
N
z
N
t
, 3 − mode unfolding
is Y
(3)
∈ R
N
z
×N
x
N
y
N
t
and 4 − mode unfolding is
Y
(4)
∈ R
N
t
×N
x
N
y
N
z
.
Definition 2: The 1-mode product of a matrix U ∈
R
J
x
×N
x
with Y ∈ R
N
x
×N
y
×N
z
×N
t
is a tensor with di-
mension (J
x
× N
y
× N
z
× N
t
) which has the entries
(Y ×
1
U)
j
x
n
y
n
z
n
t
=
∑
n
x
y
n
x
n
y
n
z
n
t
u
j
x
n
x
.
Likewise, the other n − mode multiplications are de-
fined.
Definition 3: The scalar product between two tensors
Y , Z ∈ R
N
x
×N
y
×N
z
×N
t
is
hY , Zi =
∑
n
x
∑
n
y
∑
n
z
∑
n
t
z
∗
n
x
n
y
n
z
n
t
y
n
x
n
y
n
z
n
t
,
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A Multilinear Approach
193

where
∗
represents the complex conjugate.
Whenever the scalar product between two tensors
is 0, they are orthogonal to each other. The Frobe-
nius norm of a tensor is the square root of the scalar
product of the tensor with itself, ||Y ||
F
=
p
hY , Y i.
Definition 4: The n-rank of of Y denoted by R
n
=
rank
n
(Y ), is the dimension of the vector space
spanned by the columns of the n − mode unfold-
ing. The multilinear rank for Y is represented as
rank
[]
(Y ) = [R
x
, R
y
, R
z
, R
t
] where R
x
, R
y
, R
z
, R
t
are in-
tegers between 1 and N
x
, N
y
, N
z
, N
t
, respectively.
The MLSVD decomposes the data tensor Y ∈
R
N
x
×N
y
×N
z
×N
t
into
Y = S ×
1
U
(1)
×
2
U
(2)
×
3
U
(3)
×
4
U
(4)
,
where S ∈ R
N
x
×N
y
×N
z
×N
t
is an all-orthogonal tensor.
An all-orthogonal tensor has the following properties.
Let a subtensor be defined by fixing an n − mode in-
dex of S. For example let the subtensor created by
fixing the first mode value n
x
∈ [1, ..., N
x
] to α be
defined as S
n
x
=α
. This subtensor S
n
x
=α
is orthogo-
nal to any other S
n
x
=β
for all n
x
, α and β given that
α 6= β. In addition, the frobenius norm of each subten-
sor is ordered, ||S
n
x
=1
||
F
≥ ||S
n
x
=2
||
F
... ≥ ||S
n
x
=N
x
||
F
.
The matrices U
(1)
∈ R
N
x
×N
x
, U
(2)
∈ R
N
y
×N
y
, U
(3)
∈
R
N
z
×N
z
and U
(4)
∈ R
N
t
×N
t
are unitary.
3 SIGNAL MODEL
In nearly all commercial scanners the recordings are
logarithmically compressed to deal with the large dy-
namic ranges. Here we model the logarithmically
compressed DCEUS recordings as the addition of the
original signal and the noise,
Y = G + E , (1)
where G ∈ R
N
x
×N
y
×N
z
×N
t
stands for the original sig-
nal, Y ∈ R
N
x
×N
y
×N
z
×N
t
stands for the received signal
and E ∈ R
N
x
×N
y
×N
z
×N
t
stands for the noise. When
only multiplicative noise is considered, and the log-
arithmic compression is applied, E stands for the
speckle noise (Barrois et al., 2013). After the loga-
rithmic compression, speckle noise is shown to obey
the Fisher-Tippet distribution, which can be approx-
imated as a white Gaussian noise with outliers that
has a fixed variance (Michailovich and Tannenbaum,
2006). There can be several other noise sources in
practical scenarios, such as the motion artifacts due
to the urologist’s probe handling and the patient’s
breathing.
The time-intensity curves G are assumed to follow
the modified local density random walk (mLDRW)
model (Kuenen et al., 2011), which is given as
g
xyzt
= α
xyz
r
κ
xyz
2π(t −t
0
)
e
−
κ
xyz
(t −t
0
−µ
xyz
)
2
2(t −t
0
)
. (2)
Here, κ is the local dispersion-related parameter inde-
pendent of the injection site’s distance. For low values
of dispersion, a symmetric curve and a high κ value
are observed. This is expected to represent malignant
regions (Mischi et al., 2009) (Kuenen et al., 2011).
On the other hand, a low κ is expected to represent
the benign regions.
4 PROPOSED ALGORITHM
The data tensor Y is a 4D DCEUS recording where
the first three are the spatial dimensions in the carte-
sian domain, and the fourth is the temporal dimen-
sion. We recover G by truncating Y with ranks
[R
x
, R
y
, R
z
, R
t
] that obey the condition 1 < R
x
< N
x
,
1 < R
y
< N
y
, 1 < R
z
< N
z
and 1 < R
t
< N
t
. This
assumption is expected to hold since the movement
of the microbubbles is bounded by the spatial loca-
tions of the vascular architecture inside the prostate,
and their temporal characteristics are a latent variable
of indicator dilution models. In addition, we assume
that the noise is independent of the signal itself. With
these assumptions, the problem at hand transforms
into a tensor rank estimation problem, where the rank
that defines the signal subspace will be estimated, and
the original signal will be recovered.
The multilinear truncation is done on each n −
mode unfolding separately. This can be described as
Y
(1)
= U
(1)
Σ
(1)
V
(1)
T
= Σ
(1)
×
1
U
(1)
×
2
V
(1)
Y
(2)
= U
(2)
Σ
(2)
V
(2)
T
= Σ
(2)
×
1
U
(2)
×
2
V
(2)
Y
(3)
= U
(3)
Σ
(3)
V
(3)
T
= Σ
(3)
×
1
U
(3)
×
2
V
(3)
Y
(4)
= U
(4)
Σ
(4)
V
(4)
T
= Σ
(4)
×
1
U
(4)
×
2
V
(4)
Truncate each singular vector by first R
i
< N
i
for
i ∈ [x, y, z, t]. This can be described by the operation
Y
(i)
=
h
¯
U
(i)
˜
U
(i)
i
"
Σ
(i)
R
i
0
0 Σ
(i)
N
i
−R
i
#"
¯
V
(i)
T
˜
V
(i)
T
#
(3)
where
¯
U
(i)
is the column-wise stacking of first R
i
vec-
tors representing the eigenvectors of the highest sin-
gular values,
˜
U
(i)
is the column-wise stacking of the
N
i
− R
i
columns that represent the eigenvectors of the
rest of the singular values,
¯
V
(i)
T
and
˜
V
(i)
T
are defined
in a similar way but represent the right eigenvectors
of the ith unfolding of Y .
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
194

The multilinear ranks r ank
[]
G are estimated us-
ing the SCORE algorithm proposed by (Yokota et al.,
2016). The truncation is done by projecting each
mode to the column subspace represented by the esti-
mated rank
ˆ
G = Y ×
1
¯
U
(1)
¯
U
(1)
T
×
2
¯
U
(2)
¯
U
(2)
T
×
3
¯
U
(3)
¯
U
(3)
T
(4)
×
4
¯
U
(4)
¯
U
(4)
T
,
where
ˆ
G represent the estimate of the TICs.
5 EXPERIMENTAL RESULTS
Two simulations and an in-vivo analysis are reported
in section 5.1 and section 5.2, respectively. In sec-
tion 5.1, a theoretical analysis is done to compare
the performances of SVD and MLSVD for two noise
scenarios. The TICs are generated according to the
model described at (2). The speckle noise is added for
the best-case scenario. Additionally, motion artifacts
and white Gaussian noise are added for the worst-
case scenario. The resulting signals are logarithmi-
cally compressed and then truncated using SVD and
MLSVD. The ranks are estimated using two methods
that are abbreviated as mlsvd min and mlsvd score.
In the former, the rank that gives the least MSE is
chosen and in the latter, the SCORE algorithm pro-
posed by (Yokota et al., 2016) is used. The estimated
signals are fit using the mLDRW model given at (2)
using the algorithm described in (Kuenen et al., 2011)
after reverting the logarithmic compression. For each
algorithm, the mean squared error is calculated by
MSE =
1
N
x
N
y
N
z
N
t
N
x
∑
x=1
N
y
∑
y=1
N
z
∑
z=1
N
t
∑
t=1
( ˆg
xyzt
− g
xyzt
)
2
. (5)
5.1 Simulation
We consider a signal G ∈ R
10×10×10×30
which holds
TICs that obey the mLDRW model as described at
(2). The voxel size is 0.75 mm, and the time step is
4 seconds. The simulated region holds three different
TICs, which have the parameters that are commonly
observed in the literature (Wildeboer et al., 2020), that
is,
• T IC
1
(κ
1
, µ
1
, α
1
) = [0.5 ±0.1, 30 ± 1, 1000 ± 10] ,
• T IC
2
(κ
2
, µ
2
, α
2
) = [1 ± 0.1, 25 ± 1, 1600 ± 10] ,
• T IC
3
(κ
3
, µ
3
, α
3
) = [2 ± 0.1, 15 ± 1, 1200 ± 10] .
Let the first malignant region be defined as a
3x3x3 block located at the indices [x, y, z] = [2 : 5, 2 :
5, 2 : 5]. The second malignant region is located at
TIC
3
TIC
2
TIC
1
1 2 3 4 5 6 7 8 9 10
y
1
2
3
4
5
6
7
8
9
10
z
Figure 1: The simulation setup of 3D rectangular region
with three different TICs. The slice at x = 2 is shown. Three
different simulation areas where the dark blue (majority of
the slice) represents the first region, the light blue (left top)
rectangle represents the second region, and the yellow part
(bottom right) represents the third region.
the indices [x, y, z] = [2 : 5, 6 : 9,6 : 9], and the benign
region is the area that is not covered by the first two
blocks. A slice at x = 2 represents these three differ-
ent regions, which is given in Figure 1. The benign
region (majority of the block) shown as dark blue is
assigned as region 1, the light blue (left top) rectangle
represents the region 2, and the yellow part (bottom
right) represents the region 3. These numbers are used
for referring to the malignant and benign regions. For
example κ
1
will refer to the κ values inside the re-
gion 1, κ
2
for region 2 and κ
3
for region 3. The other
parameters are represented in the same fashion. The
true rank of the generated signal G in this setup is
rank
[]
G = [2, 3, 3, 3].
Two noise scenarios are simulated. For Scenario
1, the TICs are noised with Rayleigh-shaped multi-
plicative noise. For Scenario 2, a variety of noise
sources have been added. In addition to the multi-
plicative noise, we have added a sinusoidal breath-
ing artifact with an amplitude of 0.5 mm and a fre-
quency of 0.2 Hz, a random-walk displacement that
has the maximum translation of 0.0125 mm at each
step to simulate the probe-handling of the urologist,
white Gaussian noise with 4 dB SNR where the SNR
is calculated with respect to the averaged TICs. For
each case, the error measures given at (5) is calcu-
lated and averaged across 100 Monte Carlo simula-
tions. A plot of MSE can be observed in Figure 2 and
Figure 4 for Scenario 1 and Scenario 2, respectively.
Plot (a) show the MSE over 100 iterations when the
highest 1 to 7 principal components are used for trun-
cation. Since MLSVD does not share the same x axis,
the MSE is drawn as a straight line on the same plot.
The line with the circle markers is the average MSE
over 100 iterations when the best performing trunca-
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A Multilinear Approach
195
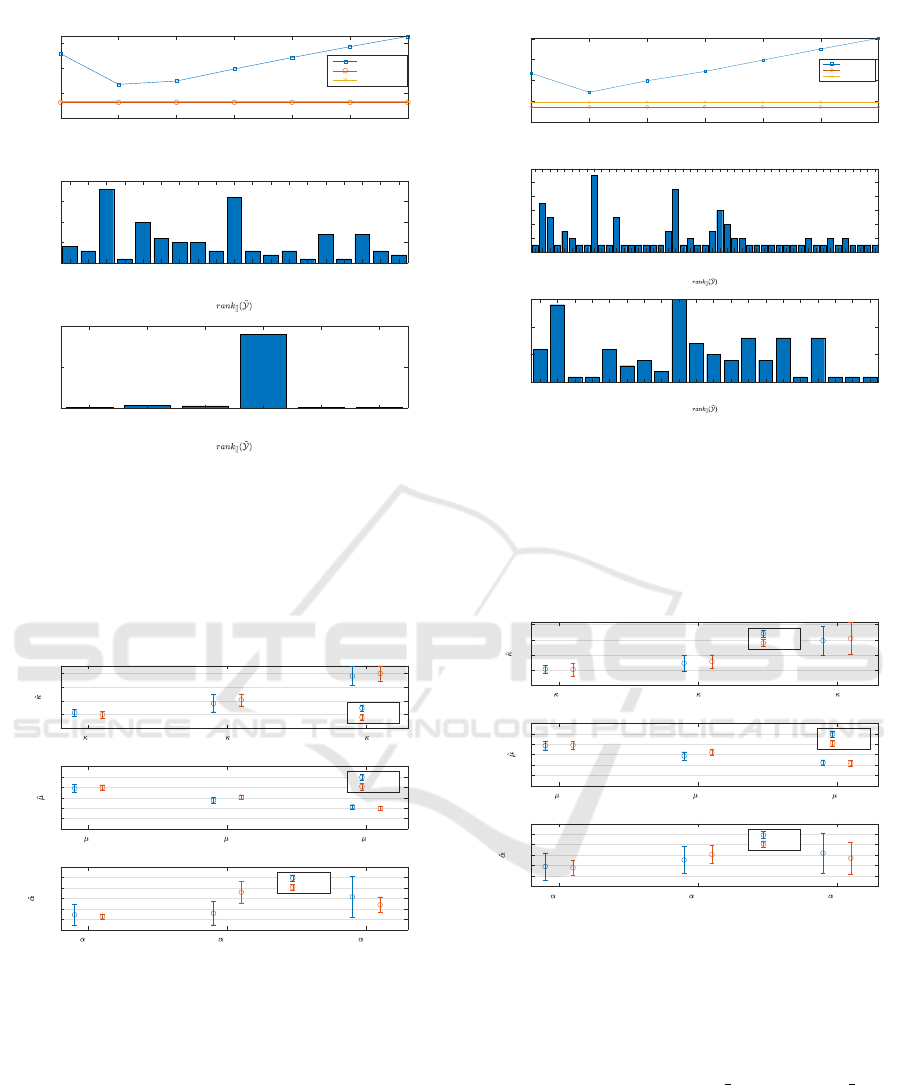
1 2 3 4 5 6 7
principal components
0
20
40
60
MSE
(a)
svd
mlsvd_min
mlsvd_score
[2,10,3,3]
[2,3,10,3]
[2,3,3,3]
[2,3,3,6]
[2,3,4,3]
[2,3,5,3]
[2,3,6,3]
[2,3,7,3]
[2,3,9,3]
[2,4,3,3]
[2,4,4,3]
[2,4,5,3]
[2,4,6,3]
[2,4,7,3]
[2,5,3,3]
[2,5,5,3]
[2,6,3,3]
[2,7,3,3]
[2,9,3,3]
0
5
10
15
20
counts
(b)
[2,2,2,3]
[2,2,3,3]
[2,3,2,3]
[2,3,3,3]
[2,3,4,3]
[2,4,3,3]
0
50
100
counts
(c)
Figure 2: The performance comparison for Scenario 1. At
(a), the MSE of SVD when several principal components
are used to truncate is shown along with two lines that show
the best performing MLSVD error over 100 iterations and
the performance of MLSVD after the ranks are estimated
with SCORE algorithm. At (b), the histogram of the ranks
that gives the least MSE at each simulation is shown. At
(c), the histogram of the ranks that are estimated with the
SCORE algorithm is shown.
TIC
1
(
1
= 0.5) TIC
2
(
2
= 1) TIC
3
(
3
= 2)
TIC curve types
0
0.5
1
1.5
2
svd
mlsvd
TIC
1
(
1
= 30) TIC
2
(
2
=25) TIC
3
(
3
= 15)
TIC curve types
10
15
20
25
30
35
40
svd
mlsvd
TIC
1
(
1
= 1000) TIC
2
(
2
= 1600) TIC
3
(
3
= 1200)
TIC curve types
800
1000
1200
1400
1600
1800
2000
svd
mlsvd
Figure 3: Plot that shows the mean and the standard devi-
ation of the ˆµ,
ˆ
κ and
ˆ
α for three different TICs in Scenario
1. The circles represents the mean values whereas, the error
bars represents the standard deviation. The mean and the
standard deviation are calculated over 100 iterations.
tion is applied, and the line with cross markers is the
MSE when the ranks are estimated using the SCORE
algorithm. Plot (b) and (c) represent the histogram
of the ranks that give the least MSE and the rank esti-
mated by the SCORE algorithm, respectively. The es-
timated κ and µ parameters are shown in Figure 3 and
Figure 5, respectively. In 87 percent of the cases, the
correct rank is estimated by the SCORE algorithm,
1 2 3 4 5 6 7
principal components
20
40
60
80
100
MSE
(a)
svd
mlsvd_min
mlsvd_score
[2,10,3,3]
[2,3,3,4]
[2,3,4,4]
[2,3,4,5]
[2,3,5,3]
[2,3,5,4]
[2,3,6,3]
[2,3,7,3]
[2,4,3,4]
[2,4,3,5]
[2,4,4,3]
[2,4,4,4]
[2,4,4,5]
[2,4,5,4]
[2,4,6,3]
[2,4,6,4]
[2,4,9,3]
[2,5,2,4]
[2,5,3,3]
[2,5,3,4]
[2,5,3,5]
[2,5,4,4]
[2,5,4,5]
[2,5,5,4]
[2,6,3,3]
[2,6,3,4]
[2,7,3,4]
[2,8,3,3]
[2,8,3,4]
[2,8,5,2]
[2,9,3,3]
[2,9,3,4]
[2,9,4,3]
[3,3,6,2]
[3,5,3,2]
[3,5,4,2]
[3,6,3,2]
[3,7,3,2]
[3,7,3,3]
[3,8,2,2]
[3,8,3,2]
[3,9,3,2]
[4,3,3,2]
[4,4,3,2]
[4,4,4,2]
[7,3,3,2]
[8,3,3,2]
0
2
4
6
8
10
12
counts
(b)
[3,2,2,3]
[3,2,2,4]
[3,2,2,5]
[3,2,3,3]
[3,2,3,4]
[3,3,2,3]
[3,3,2,4]
[3,3,3,3]
[3,3,3,4]
[4,2,2,3]
[4,2,2,4]
[4,2,3,3]
[4,2,3,4]
[4,3,2,3]
[4,3,2,4]
[4,3,3,3]
[4,3,3,4]
[5,2,2,4]
[5,3,2,4]
[5,3,3,2]
0
5
10
15
counts
(c)
Figure 4: The performance comparison for Scenario 2. At
(a), the MSE of SVD when several principal components
are used to truncate is shown along with two lines that show
the best performing MLSVD error over 100 iterations and
the performance of MLSVD after the ranks are estimated
with SCORE algorithm. At (b) the histogram of the ranks
that gives the least MSE at each simulation are shown. At
(c) the histogram of the ranks estimated with SCORE algo-
rithm shown.
TIC
1
(
1
= 0.5) TIC
2
(
2
= 1) TIC
3
(
3
= 2)
TIC curve types
0
0.5
1
1.5
2
svd
mlsvd
TIC
1
(
1
= 30) TIC
2
(
2
= 25) TIC
3
(
3
= 15)
TIC curve types
10
15
20
25
30
35
40
svd
mlsvd
TIC
1
(
1
= 1000) TIC
2
(
2
= 1600) TIC
3
(
3
= 1200)
TIC curve types
800
1000
1200
1400
1600
1800
2000
svd
mlsvd
Figure 5: Plot that shows the mean and the standard devi-
ation of the ˆµ,
ˆ
κ and
ˆ
α for three different TICs at Scenario
2. The circles represents the mean values whereas, the error
bars represents the standard deviation. The mean and the
standard deviation are calculated over 100 iterations.
which can be seen at plot (c) of Figure 2. For this
case, the performance of mlsvd min and mlsvd score
overlap suggesting that the SCORE algorithm gives
the least mse over 100 iterations. Although a perfor-
mance improvement over SVD is observed in Figure
4, the ranks are not estimated correctly when a variety
of noise sources are added. The reasons are discussed
at Section 6.
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
196
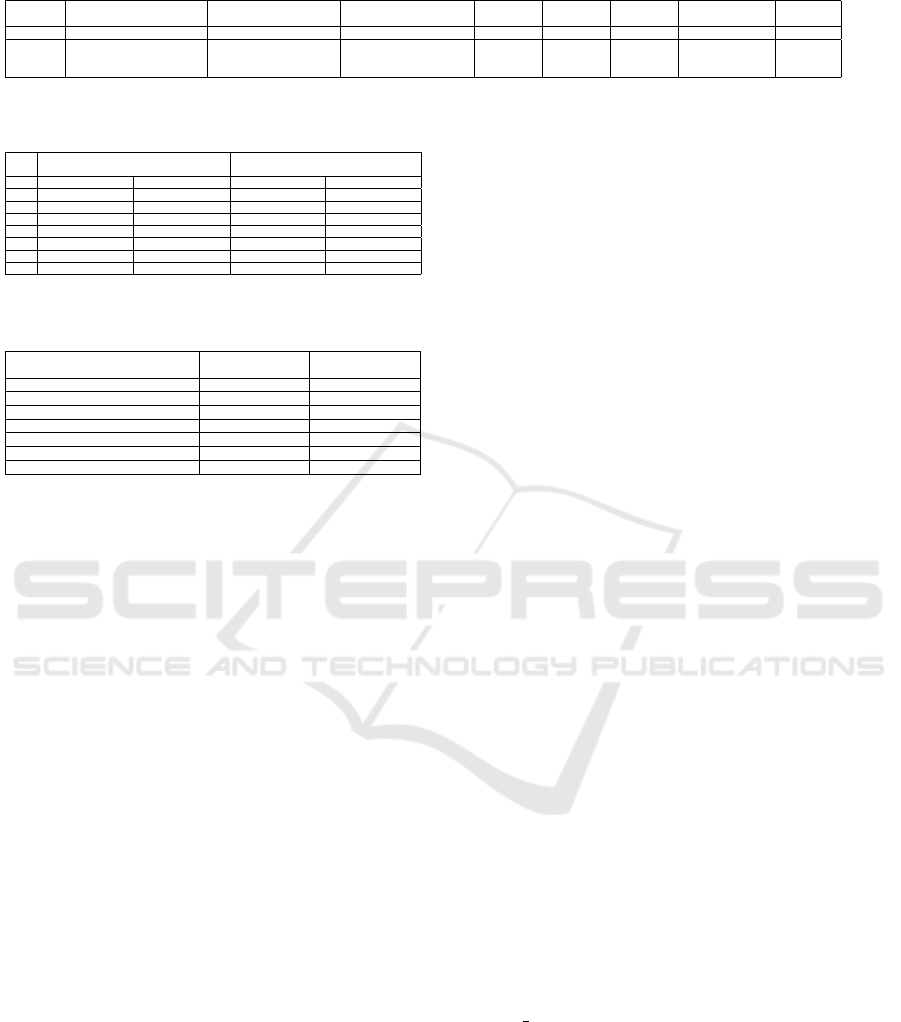
Table 1: The summary of simulation scenarios.
T IC
1
(κ
1
, µ
1
, α
1
) T IC
2
(κ
2
, µ
2
, α
2
) T IC
3
(κ
3
, µ
3
, α
3
)
Number of
Voxels TIC
1
Number of
Voxels TIC
2
Number of
Voxels TIC
3
Noise Types
Number of
Simulations
Scenario 1 (0.5 ± 0.1,30 ± 1, 1000 ± 10) (1 ± 0.1, 25 ± 1, 1600 ± 10) (2 ± 0.1, 15 ± 1, 1200 ± 10) 872 64 64 Multiplicative 100
Scenario 2 (0.5 ± 0.1,30 ± 1, 1000 ± 10) (1 ± 0.1, 25 ± 1, 1600 ± 10) (2 ± 0.1, 15 ± 1, 1200 ± 10) 872 64 64
Multiplicative,
Breathing, Motion,
WGN
100
Table 2: The estimated model fit parameters across all pa-
tients.
SVD
MLSVD-SCORE
ρ = 10e − 4
Benign Malignant Benign Malignant
κ 0.51(σ = 0.35) 0.56(σ = 0.33) 0.39(σ = 0.18) 0.48(σ = 0.22)
PV 94.73(σ = 29.57) 104.05(σ = 27.53) 87.87(σ = 26.33) 98.49(σ = 24.03)
PT 33.76(σ = 9.57) 30.00(σ = 8.09) 32.56(σ = 6.95) 29.24(σ = 5.62)
AT 15.12(σ = 4.96) 13.72(σ = 4.23) 14.32(σ = 3.44) 13.41(σ = 3.11)
WIT 18.64(σ = 9.41) 16.28(σ = 7.44) 18.24(σ = 5.91) 15.84(σ = 4.08)
µ 35.28(σ = 11.48) 31.31(σ = 9.57) 34.29(σ = 8.31) 30.58(σ = 6.24)
α 172.99(σ = 126.38) 184.39(σ = 123.05) 143.78, (σ = 77.96) 154.09, (σ = 68.92)
Table 3: The classification performance across all the pa-
tients.
SVD
MLSVD-SCORE
(ρ = 10e − 4)
[κ
sens
, κ
spec
, κ
AUC
,threshold] [0.54,0.53,0.57,0.51] [0.48,0.65,0.63,0.45]
[PI
sens
, PI
spec
, PI
AUC
,threshold] [0.60,0.54,0.61,99] [0.63,0.57,0.64,93]
[PT
sens
, PT
spec
, PT
AUC
,threshold] [0.57,0.62,0.61,31] [0.59,0.65,0.63,30]
[α
sens
, α
spec
, α
AUC
,threshold] [0.54,0.51,0.55,150] [0.46,0.64,0.58,160]
[W IR
sens
,W IR
spec
,W IR
AUC
,threshold] [0.58,0.59,0.63,6.6] [0.62,0.59,0.66,5.7]
[AT
sens
, AT
spec
, AT
AUC
,threshold] [0.65,0.58,0.59,15] [0.72,0.59,0.58,13.8]
[W IT
sens
,W IT
spec
,W IT
AUC
,threshold] [0.58,0.57,0.56,16] [0.59,0.58,0.62,16.5]
5.2 In-vivo Analysis
The recording of 6 patients was acquired from the
Second Affiliated Hospital of Zheijang University
(Hangzhou, Zheijang, China). Written consent was
obtained. A 2.4-mL bolus of SonoVue
®
was in-
travenously injected, and a 4D recording in con-
trast mode was obtained with a LOGIQ E9 scanner
equipped with a RIC5-9-D endocavitary transducer
driven at 4 MHz. The volume rate was fixed to 0.25
Hz by setting the image quality to low, and the dis-
ruption of microbubbles was avoided by fixing the
mechanical index to 0.1. The patients went through
prostatectomy after the recording. The prostate was
sliced with 4-mm thickness, and for each slice, an
annotation was made by the pathologist. The an-
notations were registered back to the domain of the
recording, and the ground truth was obtained. Only
two out of the six patients had significant lesions with
Grade Group > 3. A region of interest is selected in
the benign and malignant regions that are reasonably
close to the true annotations. There are around 25000
voxels, of which 18000 are malignant.
The spatial resolution of the recording is regular-
ized in space, and the data is downsampled by a fac-
tor of 3 as described by (Schalk et al., 2015b). The
MLSVD is applied, and the signal is truncated us-
ing the ranks estimated by the SCORE (Yokota et al.,
2016) algorithm where ρ = 10e − 4 is selected. The
mLDRW model is fit as described at (Kuenen et al.,
2011), and the perfusion and dispersion parameters
are extracted. The results can be seen in Table 2 and
Table 3. In the former, the mean and the standard de-
viation of the features are shown. In the latter, the
sensitivity, specificity, and area under the receiver op-
erating characteristic curves are shown. The classifi-
cation is done by determining the point in the receiver
operating characteristic curve that is the closest to the
upper left corner (sensitivity and specificity equal to
1) in Euclidean distance.
6 DISCUSSION
In this paper, the DCEUS sequences are denoised us-
ing SVD and MLSVD, and their performances are
evaluated based on the quality of the TIC model-
ing. In section 5.1, two simulations are reported that
employ the commonly observed noises in DCEUS
recordings. The first simulation represents the best-
case scenario where only speckle-noise exists. The
second simulation represents the worst-case scenario
where additive WGN and motion exist apart from
the speckle noise. MLSVD mostly performs better
at capturing the signal characteristics in both cases,
providing a better estimate of dispersion and perfu-
sion parameters. Both methods perform worse in the
worst-case scenario and fail to estimate high κ values
precisely. The low SNR might explain the decrease in
performance.
The speckle noise can be modeled as a WGN
with outliers (Michailovich and Tannenbaum, 2006),
which results in the correct estimation of the rank for
Scenario 1. The actual rank of the signal is [2, 3, 3, 3],
which is the same as the rank that is estimated the
majority of the time. For this case, we can see in Fig-
ure 2 that the MSE of the best performing rank and
the estimated rank overlap. In the cases where the
rank was incorrectly estimated, the error is found to
be within 15% of the MSE represented with the line
mlsvd score. The estimation of the correct rank fails
for Scenario 2. Adding the other noise sources vio-
lates the WGN assumption and causes the rank esti-
mator to perform poorly. This suggests that in a prac-
tical scenario where the recording suffers from var-
ious noise sources apart from the speckle noise, the
proposed algorithm might not be suitable. In the fig-
ures Figure 2 and Figure 4, it can be observed that the
best performing rank is not necessarily the true rank
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A Multilinear Approach
197

of the data. The subfigures labeled with (b) in both
plots suggest that the noise in some cases might oc-
cupy the highest ranks. For this reason, a higher rank
might have provided a lower MSE.
Better classification performance of MLSVD over
SVD is observed at Table 3. The improvement is more
significant when a subset of the patients is considered.
This might be due to the insignificant malignant vox-
els of the four patients that are investigated. The sig-
nificant malignant lesions are expected to have a more
distinct characteristic than benign tissue. This behav-
ior is observed for two of the patients, who have sig-
nificant lesions. The AUC is found to be greater than
0.8 for these two patients whereas, a similar perfor-
mance has not been observed for the others.
7 CONCLUSION
In this paper, we proposed a denoising algorithm for
detecting prostate cancer from 4D DCEUS record-
ings. Previously, SVD was proposed to denoise the
single-dimensional TICs that suffer from low SNR.
Here, we have retained the volumetric information
by considering the tensor format of the recording
and introduced an algorithm based on MLSVD and
SCORE. The proposed algorithm is shown to outper-
form SVD in simulation and in in-vivo experiments.
The in-vivo results have poor classification perfor-
mance, which can be due to the insignificant lesions
found in four of the six patients that have undergone
prostatectomy. In future work, we plan to use a larger
dataset and include other features used in the detec-
tion of angiogenesis.
ACKNOWLEDGEMENTS
This project is funded in part by Holland High Tech
with a PPS supplement for research and development
in the Topsector HTSM. We would like to acknowl-
edge Prof. Pintong Huang for carrying out the clini-
cal trials at the Second Affiliated Hospital of Zheijang
University (Hangzhou, Zheijang, China).
REFERENCES
Bar-Zion, A. D., Tremblay-Darveau, C., Yin, M., Adam,
D., and Foster, F. S. (2015). Denoising of contrast-
enhanced ultrasound cine sequences based on a mul-
tiplicative model. IEEE Transactions on Biomedical
Engineering, 62(8):1969–1980.
Barrois, G., Coron, A., Payen, T., Dizeux, A., and Bridal,
L. (2013). A multiplicative model for improving
microvascular flow estimation in dynamic contrast-
enhanced ultrasound (dce-us): theory and experimen-
tal validation. IEEE transactions on ultrasonics, fer-
roelectrics, and frequency control, 60(11):2284–2294.
Bjaerum, S., Torp, H., and Kristoffersen, K. (2002). Clutter
filter design for ultrasound color flow imaging. IEEE
Transactions on Ultrasonics, Ferroelectrics, and Fre-
quency Control, 49(2):204–216.
De Lathauwer, L., De Moor, B., and Vandewalle, J.
(2000). A multilinear singular value decomposition.
SIAM journal on Matrix Analysis and Applications,
21(4):1253–1278.
Delorme, S. and Knopp, M. (1998). Non-invasive vascu-
lar imaging: assessing tumour vascularity. European
radiology, 8(4):517–527.
Eckersley, R. J., Sedelaar, J. M., Blomley, M. J., Wijk-
stra, H., deSouza, N. M., Cosgrove, D. O., and de la
Rosette, J. J. (2002). Quantitative microbubble en-
hanced transrectal ultrasound as a tool for monitor-
ing hormonal treatment of prostate carcinoma. The
Prostate, 51(4):256–267.
Elie, N., Kaliski, A., P
´
eronneau, P., Opolon, P., Roche, A.,
and Lassau, N. (2007). Methodology for quantify-
ing interactions between perfusion evaluated by dce-
us and hypoxia throughout tumor growth. Ultrasound
in medicine & biology, 33(4):549–560.
Elwenspoek, M. M. C., Sheppard, A. L., McInnes, M. D. F.,
Merriel, S. W. D., Rowe, E. W. J., Bryant, R. J.,
Donovan, J. L., and Whiting, P. (2019). Compari-
son of Multiparametric Magnetic Resonance Imaging
and Targeted Biopsy With Systematic Biopsy Alone
for the Diagnosis of Prostate Cancer: A Systematic
Review and Meta-analysis. JAMA Network Open,
2(8):e198427–e198427.
Gillies, R. J., Schomack, P. A., Secomb, T. W., and Raghu-
nand, N. (1999). Causes and effects of heterogeneous
perfusion in tumors. Neoplasia, 1(3):197–207.
Joel, T. and Sivakumar, R. (2018). An extensive review
on despeckling of medical ultrasound images using
various transformation techniques. Applied Acoustics,
138:18–27.
Kuenen, M. P. J., Mischi, M., and Wijkstra, H. (2011).
Contrast-ultrasound diffusion imaging for localization
of prostate cancer. IEEE Transactions on Medical
Imaging, 30(8):1493–1502.
Liu, G., Wu, S., and Huang, L. (2020). Contrast-enhanced
ultrasound evaluation of the prostate before transrectal
ultrasound-guided biopsy can improve diagnostic sen-
sitivity: A stard-compliant article. Medicine, 99(19).
Loeb, S., Vellekoop, A., Ahmed, H. U., Catto, J., Ember-
ton, M., Nam, R., Rosario, D. J., Scattoni, V., and
Lotan, Y. (2013). Systematic review of complications
of prostate biopsy. European Urology, 64(6):876–892.
Michailovich, O. and Tannenbaum, A. (2006). Despeckling
of medical ultrasound images. IEEE Transactions on
Ultrasonics, Ferroelectrics, and Frequency Control,
53(1):64–78.
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
198

Mischi, M., Kalker, A., and Korsten, H. (2003). Video-
densitometric methods for cardiac output measure-
ments. EURASIP Journal on Applied Signal Process-
ing, 2003(5):479–489.
Mischi, M., Kuenen, M., Wijkstra, H., Hendrikx, A., and
Korsten, H. (2009). Prostate cancer localization by
contrast-ultrasound diffusion imaging. In 2009 IEEE
International Ultrasonics Symposium, pages 283–
286.
Mottet, N., Bellmunt, J., Bolla, M., Briers, E., Cumber-
batch, M. G., De Santis, M., Fossati, N., Gross, T.,
Henry, A. M., Joniau, S., Lam, T. B., Mason, M. D.,
Matveev, V. B., Moldovan, P. C., van den Bergh, R. C.,
Van den Broeck, T., van der Poel, H. G., van der
Kwast, T. H., Rouvi
`
ere, O., Schoots, I. G., Wiegel, T.,
and Cornford, P. (2017). Eau-estro-siog guidelines on
prostate cancer. part 1: Screening, diagnosis, and lo-
cal treatment with curative intent. European Urology,
71(4):618–629.
Ozgun, K. and Byram, B. (2020). A channel domain higher-
order svd clutter rejection filter for small vessel ultra-
sound imaging. In 2020 IEEE International Ultrason-
ics Symposium (IUS), pages 1–4.
Russo, G., Mischi, M., Scheepens, W., De la Rosette, J. J.,
and Wijkstra, H. (2012). Angiogenesis in prostate can-
cer: onset, progression and imaging. BJU Interna-
tional, 110(11c):E794–E808.
Schalk, S. G., Demi, L., Smeenge, M., Mills, D. M., Wal-
lace, K. D., de la Rosette, J. J. M. C. H., Wijkstra, H.,
and Mischi, M. (2015a). 4-d spatiotemporal analy-
sis of ultrasound contrast agent dispersion for prostate
cancer localization: a feasibility study. IEEE Trans-
actions on Ultrasonics, Ferroelectrics, and Frequency
Control, 62(5):839–851.
Schalk, S. G., Demi, L., Smeenge, M., Mills, D. M., Wal-
lace, K. D., de la Rosette, J. J. M. C. H., Wijkstra, H.,
and Mischi, M. (2015b). 4-d spatiotemporal analy-
sis of ultrasound contrast agent dispersion for prostate
cancer localization: a feasibility study. IEEE Trans-
actions on Ultrasonics, Ferroelectrics, and Frequency
Control, 62(5):839–851.
Seitz, M., Shukla-Dave, A., Bjartell, A., Touijer, K., Scia-
rra, A., Bastian, P. J., Stief, C., Hricak, H., and Graser,
A. (2009). Functional magnetic resonance imaging in
prostate cancer. European Urology, 55(4):801–814.
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A.
(2021). Cancer statistics, 2021. CA: A Cancer Journal
for Clinicians, 71(1):7–33.
Strouthos, C., Lampaskis, M., Sboros, V., McNeilly, A., and
Averkiou, M. (2010). Indicator dilution models for the
quantification of microvascular blood flow with bolus
administration of ultrasound contrast agents. IEEE
transactions on ultrasonics, ferroelectrics, and fre-
quency control, 57(6):1296–1310.
Ukimura, O., Coleman, J. A., de la Taille, A., Emberton,
M., Epstein, J. I., Freedland, S. J., Giannarini, G., Ki-
bel, A. S., Montironi, R., Ploussard, G., Roobol, M. J.,
Scattoni, V., and Jones, J. S. (2013). Contemporary
role of systematic prostate biopsies: Indications, tech-
niques, and implications for patient care. European
Urology, 63(2):214–230.
Wildeboer, R., Van Sloun, R., Mannaerts, C., Van der Lin-
den, J., Huang, P., Wijkstral, H., and Mischi, M.
(2018a). Probabilistic 3d contrast-ultrasound trac-
tography based on a a convective-dispersion finite-
element scheme. In 2018 IEEE International Ultra-
sonics Symposium (IUS), pages 1–9.
Wildeboer, R. R., Sammali, F., van Sloun, R. J. G., Huang,
Y., Chen, P., Bruce, M., Rabotti, C., Shulepov, S., Sa-
lomon, G., Schoot, B. C., Wijkstra, H., and Mischi, M.
(2020). Blind source separation for clutter and noise
suppression in ultrasound imaging: Review for differ-
ent applications. IEEE Transactions on Ultrasonics,
Ferroelectrics, and Frequency Control, 67(8):1497–
1512.
Wildeboer, R. R., van Sloun, R. J., Huang, P., Wijkstra,
H., and Mischi, M. (2019). 3-d multi-parametric
contrast-enhanced ultrasound for the prediction of
prostate cancer. Ultrasound in medicine & biology,
45(10):2713–2724.
Wildeboer, R. R., Van Sloun, R. J. G., Schalk, S. G.,
Mannaerts, C. K., Van Der Linden, J. C., Huang, P.,
Wijkstra, H., and Mischi, M. (2018b). Convective-
dispersion modeling in 3d contrast-ultrasound imag-
ing for the localization of prostate cancer. IEEE Trans-
actions on Medical Imaging, 37(12):2593–2602.
Yokota, T., Lee, N., and Cichocki, A. (2016). Robust multi-
linear tensor rank estimation using higher order sin-
gular value decomposition and information criteria.
IEEE Transactions on Signal Processing, 65(5):1196–
1206.
Zhu, Y., Kim, M., Hoerig, C., and Insana, M. F. (2020). Ex-
perimental validation of perfusion imaging with hosvd
clutter filters. IEEE Transactions on Ultrasonics, Fer-
roelectrics, and Frequency Control, 67(9):1830–1838.
Denoising of Dynamic Contrast-enhanced Ultrasound Sequences: A Multilinear Approach
199
