
Exploring Classification in Open and Closed Eyes EEG Data for
People with Cognitive Disorders
Ioanna Chouvarda
1a
, Lampros Mpaltadoros
1b
, Ioanna Boutziona
1
, George Nikolaos Tsakonas
1
,
Magda Tsolaki
2c
and Konstantinos Diamantaras
3d
1
Lab of Computing Medical Informatics and Biomedical Imaging Technologies, School of Medicine,
Aristotle University of Thessaloniki, Greece
2
Greek Alzheimer Association, Thessaloniki, Macedonia, Greece
3
Department of Information and Electronic Engineering, International Hellenic University, Thessaloniki, Greece
Keywords: Alzheimer’s Disease, Mild Cognitive Impairment, EEG, Signal Processing, Machine Learning.
Abstract: Cognitive disorders, including Alzheimer’s Disease (AD), are health issues concerning all society. The
evolution of technology and Artificial Intelligence (AI)/ Machine Learning (ML) in the health domain
promises an earlier and more accurate diagnosis for Alzheimer’s disease and Dementia. In this study, we
examine Healthy patients and patients with AD and Mild Cognitive Impairment (MCI), often a prior step of
AD. With the use of EEG, we collect data from their brain activity. After a basic processing step, kernel PCA
is applied as a dimensionality reduction method using segments of the multichannel signal, and the
transformation output is employed as input for the predictive model. Machine learning functions are used to
classify data correctly into Healthy, AD, MCI classes, and a postprocessing step allows for classification at
the patient level. The results show that the algorithm can predict with an accuracy of 90 percent and more in
total, AD or MCI patients vs. Healthy patients.
1 INTRODUCTION
The aging population is increasing at an alarming
rate. The prevalence of diseases more frequent in
older adults like Dementia is therefore increasing.
Because of the heterogeneity of clinical presentation
and complexity of disease neuropathology, dementia
classification remains controversial (Raz et al., 2016).
Current research also focused on investigating
patients with mild cognitive impairment that will
evolve to Alzheimer’s disease (Dallora et al., 2017).
An early characterisation of MCI, especially
progressing MCI, may help timely interventions and
slow disease progression.
There are many studies concerning Dementia and
AD. AD is the most common type of Dementia. The
difference between Dementia and AD is that AD has
a higher severity of EEG abnormalities (Kulkarni &
Bairagi, 2014). MCI on the other hand, is also
characterized from memory loss but is an early stage
a
https://orcid.org/0000-0001-8915-6658
b
https://orcid.org/0000-0001-8652-7628
c
https://orcid.org/0000-0002-2072-8010
d
https://orcid.org/0000-0003-1373-4022
of Dementia and AD with no apparent symptoms.
Some MCI patients may return to the normal stage,
but a small percentage of them proceed to AD
(Amezquita-Sanchez et al., 2019).
Numerous approaches have been proposed
towards the classification of dementia patients, based
on EEG, MRI images, biomarkers, daily life tests
(Buegler M, 2020). With respect to EEG for AD and
MCI characterisation, either evoked potentials, or rest
EEG, can be of use.
EEG is a medical modality used for brain
disorders, including AD and Dementia recognition. In
most Dementia types, slow brain activity is common,
so EEG is used for diagnostic evaluation. EEG signals
are categorized based on the frequency (delta, theta,
alpha, beta and gamma) from 0.1 Hz to almost 100
Hz (Kumar & Bhuvaneswari, 2012). There are many
pieces of research concerning the detection of
Dementia, AD, and MCI. Regarding rest EEG
analysis, several approaches include feature
298
Chouvarda, I., Mpaltadoros, L., Boutziona, I., Tsakonas, G., Tsolaki, M. and Diamantaras, K.
Exploring Classification in Open and Closed Eyes EEG Data for People with Cognitive Disorders.
DOI: 10.5220/0011010100003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 4: BIOSIGNALS, pages 298-305
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

extraction, in terms of spectral, wavelet, entropy
features in specific channels, or network analysis and
connectivity features among channels, combined with
machine learning for classification. Newer studies
incorporate deep learning approaches.
In the research on early stages of AD, some
researchers used Deep Neural Networks (DNN) for
classification with Relative Power (RP) to re-
combine features from the system’s learning method,
which improved diagnosis results compared to
another NN, which contained RP features as domain
knowledge (Kim & Kim, 2018). In newer studies,
though, Multiple Signal Classification and Empirical
Wavelet Transform (MUSIC-EWT) was used to
reconstruct signals into proper EEG frequencies,
analyze them with non-linear indices to discriminate
AD from MCI patients, evaluate features with
ANOVA for feature selection and use Epoch Neural
Network (EPNN) for classification (Amezquita-
Sanchez et al., 2019). Usually, preprocessing filters
are applied to EEG signals, while Independent
Component Analysis (ICA) or Blind Source
Separation (BSS) are considered for signal
improvement, Fast Fourier Transform (FFT) or
Wavelet Transform (WT) for feature extraction and
Linear Discriminant Analysis (LDA) or Support
Vector Machine (SVM) for the classification. In
addition to FFT for feature extraction, Continuous
Wavelet Transform (CWT) can also be applied, and
for data classification, K-Nearest Neighbor (KNN)
has been used successfully (Durongbhan et al., 2019).
The current study aims to use the information
hidden in all EEG channels without selecting the most
informative ones. It is explored whether open or
closed eyes recordings, are more informative. Also,
to identify the most informative frequency zones,
high-pass and low-pass filtered versions of the signal
are used. This study explores the value of a
classification method based on Kernel PCA and
Random Forest classifier in classifying Healthy, MCI
and AD patients on the preprocessed EEG data, in the
above-mentioned schemes. Classification follows
two steps, classification of EEG segments as a first
step, and classification of patients via segment
majority voting as a second step.
2 METHODS
As a starting point, the EEG data stored in European
Data Format (EDF), which included both open and
closed eyes parts, was serialized via Python object
serialization (pickle) for more efficient data handling
of the open-eye closed-eye segments separately.
During the preprocessing of the data, the data were
segmented into multiple parts for every patient and
for every status (open eyes, closed eyes). After this
process, major artifacts were rejected via standard
deviation thresholding, and two types of filters were
used (delta-theta, and alpha-beta bands, respectively).
ML algorithms were used to study the accuracy of
different classifiers when classifying patients as MCI
patients, AD patients, or Healthy, with different
schemes, e.g., eyes closed and low-pass filtered. The
algorithms used were based on the Random Forest
(RF) Classifier as a first step classifying patient
segments and a majority voting scheme as a second
step.
These methodological steps are described in more
detail in the following subsections.
2.1 EEG Data
In this paper EEG data were collected through a set
of 21 electrodes following the 10-20 international
reference system (Fp1, Fp2, F7, F3, Fz, F4, F8, T3,
C3, Cz, C4, T4, T5, P3, Pz, P4, T6, O1 and O2) at
500Hz.
For EEG signal collection an Nihon274Kohden
Neurofax J921A system was used. Input impedance
was set to Z<10kω, and the signals were digitized
with the Neurofax EEG-12200 Ver. 01-93, and a
sampling frequency of 500Hz. The protocol used for
data acquisition of the EEG signals refers to the
resting stage that lasts for 10 minutes, from which 5
minutes the patient’s eyes are closed, and the other 5
are opened, while being seated in an upright position.
For the experiment, we used 27 AD, 22 Healthy
και 24 MCI. The data were provided from the Greek
Association of Alzheimer’s Disease and Related
Disorders, with ethical approval for use, and based on
the patient data privacy legislation, the data were
anonymized.
The EEG data collected are saved in raw EEG
EDF files. Every EDF consists of 19 EEG signal
channels. Each file contains annotations about signal
phases such as open eyes, calibration, closed eyes,
A1+A2 electrode ON. Those annotations were used
to distinguish segments into open and closed eyes and
remove irrelevant ones. Then the data were processed
and stored in pickle format for storage capacity
reasons.
Following, a preprocessing pipeline is used,
including segmentation, filtering and transformation.
Exploring Classification in Open and Closed Eyes EEG Data for People with Cognitive Disorders
299

2.2 Preprocessing
Data preprocessing includes filtering and
segmentation of data before data analysis. Filtering is
used to refine data and remove noise and artifacts.
Segmentation is a technique used in this case with
the prospect of separating data depending on the
annotations of EEG signals for the machine learning
algorithm, so that there is sufficient data to be
properly trained. The data transformation via KPCA
is applied for dimensionality reduction to provide the
classifier with a reasonable number of features
representing the information of the multichannel EEG
segments.
2.2.1 Segmentation
In this study, the recordings were segmented into
smaller chunks, allowing us to train the machine
learning algorithm with smaller data chunks and
facilitating training as our dataset was quite limited.
All open-eye and closed-eye recordings were
segmented into nonoverlapping 5 second segments.
To avoid overfitting, the maximum number of
segments per patient file was set to 45, taking into
account that the number of good quality segments per
subject varied. The segmentation into chunks also
facilitated the dimensionality reduction procedure
(see section 2.3), as it was applied in flattened
segments of length N=channels x segment_size.
2.2.2 Signal Filtering
The data segments with a much higher standard
deviation than an adapted average threshold were
automatically removed from the sample to remove
possibly significant artifacts.
In addition, in this research, we used two types of
filters. The first was a low-band Finite Impulse
Response (FIR) filter to keep delta and theta signals,
and the second filter was a high-band FIR filter for
alpha and beta signals. Those FIR filters were used to
create two different data schemes. In addition,
although channels Fp1 and Fp2 are informative, they
were removed to avoid potential artifacts, a necessary
step since segments were not manually inspected.
Thus, a scheme with 17 channels was employed. An
alternative scheme with a reduced number of
channels (10 channels in the central-temporal zone)
was also considered.
2.3 Dimensionality Reduction
In search of a method that will use the flattened
segments of length N=channels x segment_size as
sample inputs, and produce a much-reduced number
of features to be used for the classification, the typical
dimensionality reduction methods, PCA and t-SNE
(van der Maaten and Hinton; 2008) were initially
considered, with moderate results.
Kernel PCA (KPCA), an extension of PCA using
kernel methods, was adopted as a much better choice.
KPCA is used for multivariate datasets and performs
better in non-linear data. With kernel methods, KPCA
can protrude data to a higher dimension where there
are linearly separable (Wang, 2012). We chose
heuristically 160 components and radial basis
function kernel (rbf) for KPCA, with gamma
parameter to be by default 1/number of features.
These 160 KPCA components, resulting from the
transformation of each multichannel segment, are
used as classification inputs.
3 CLASSIFICATION
3.1 Segment Classification
The classification of each multichannel segment,
employing the KPCA components, employs Random
Forest (RF). RF is an ensemble method based on
Decision Trees. RF aggregates the outcome of many
individual decision trees operating as one.
In the RF classifier algorithm, we applied 80
decision trees, 5 jobs to run in parallel, balanced class
weight, and random state value=1, which is the
parameter controlling the randomness of samples
when building the trees. An SVM was considered
alternatively (Awad & Khanna, 2015), but potentially
due to the fact that the data were already transformed
via an RBF kernel, did not add better results and was
not further pursued,
The number of segments used for the
classification were 871 closed-eyes and 891 open
eyes for Healthy subjects, 2008 closed and 2096 open
eyes for MCI, 1034 closed and 1034 closed, and 1197
open eyes for AD.
3.2 Subject Classification
In order to move from segment classification to
patient classification, a hard voting scheme was
applied as a second step. The classification of each
patient’s segment contributes a vote to the
classification of a patient. The performance recall
TP/(TP+FN) was calculated among the classified
segments per patient, and a threshold >=0.6 is applied
to denote the majority and suggest whether the patient
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
300
is correctly classified based on the majority of
segments or not.
For instance, if at least 60% of a patient’s
segments were classified as AD, the patient was
categorized as an AD patient.
3.3 Training and Testing
We chose to run three different binary classifiers for
our prediction, one for AD vs. MCI, one for Healthy
vs. AD, and one for MCI vs. AD. For completeness,
a 3-class classifier was also presented.
A cross-validation strategy was followed. The
classifiers were trained 20 times, leaving in each
iteration a set of 2 patients out (one from each class)
for testing, in a leave-one-subject-out scheme. For
example, we run the classifier for Healthy and MCI
patients, and for every run, the classifier left out all
the segments of a different patient from the Healthy
and MCI class. After each run, the RF classifier
returned the training and test set accuracy and a
confusion matrix, and the two-step classification
procedure was applied for the two subjects to classify
the and Healthy or MCI. The average performance
metrics per patient were used for comparison.
4 RESULTS
This section presents the results for the binary
classifiers for AD vs MCI, Healthy vs AD and MCI
vs Healthy patients for open and closed eyes with low
and high band filter, as well as the three-class model
performance
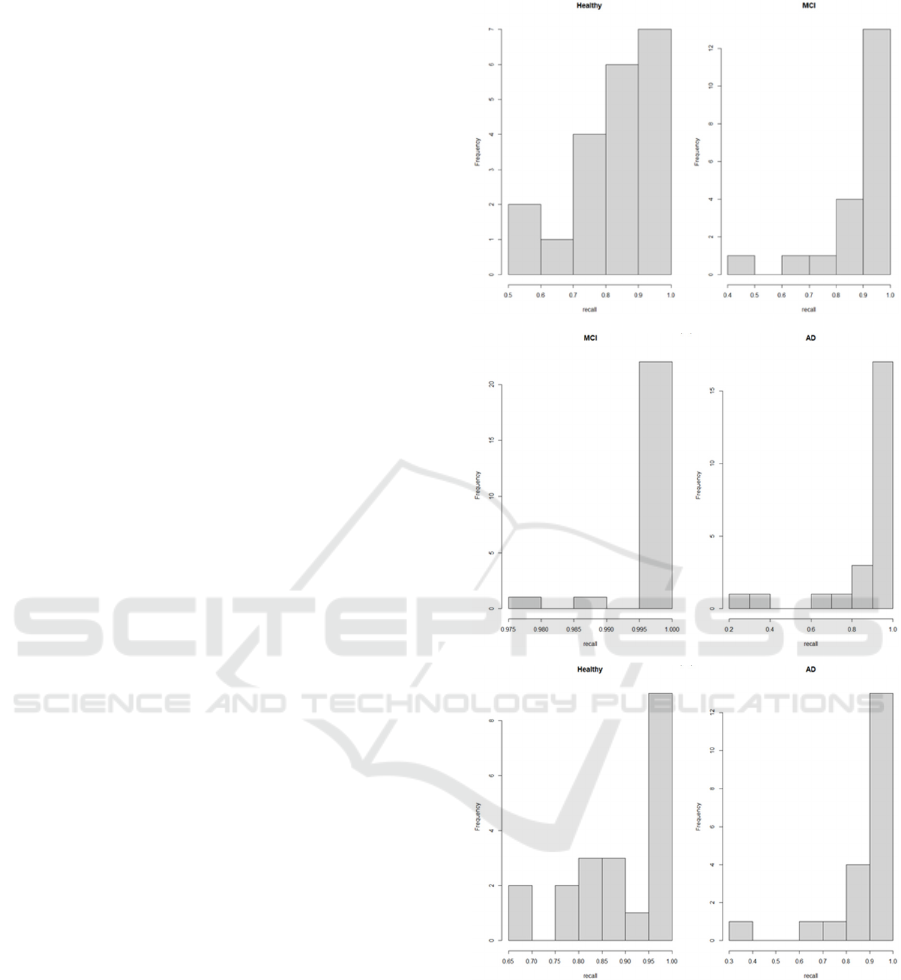
In order to illustrate the transition from the
segment-wise classification to the patient-based
classification, a histogram of the classification recall
per patient is provided (Figure 1). The recall per
patient (TP/TP+FN) shows the percentage of TP vs
FN of the classified segments per patient, and in the
hard voting scheme selected, a recall threshold >=0.6
as selected to suggest a correct patient classification.
As seen in the figures, most of the segments are well
above the 0.6 threshold in all cases. Only in Figure
1a, in the Healthy class, one can see 2 out of the 20
cases where recall is between 0-5 and 0.6, in which
cases we do not conclude with a correct subject
classification.
Table 1 presents the summarised performance
metrics in the testing set regarding the three binary
classification models, with closed eyes and low-pass
filter. In each run, all segments of the 1st class belong
to a single subject of this class that is left out for
testing,
and
the
same
stands
for
the
2nd
class.
The
(a)
(b)
(c)
Figure 1: Distribution of classification recall per patient. a)
Healthy-MCI closed eyes, low band, b) MCI-AD closed
eyes low band, c) Healthy-AD closed eyes low band.
precision and recall metrics are depicted as median
and (1
st
-3
rd
quantile), corresponding to the
percentages of correctly and falsely classified
segments per subject. The correctly classified
subjects per class are calculated based on recall >0.6
in each run.
Exploring Classification in Open and Closed Eyes EEG Data for People with Cognitive Disorders
301

Table 1: Patient Classification performance metrics for the
three binary classification models, using RF classifier and
the majority voting per patient. H stands for Healthy, M for
MCI, A for AD. #C stands for the correctly classified
subjects (correct segments > 60%).
H/
M
Precision Recall #C
H 0.97
(0.87–1)
0.84
(0.75-0.93)
18 /20
M 0.85
(0.74-0.95)
0.97
(0.86–1)
19 /20
H/
A
H 0.91
(0.85–1)
0.9
(0.81-0.97)
20/20
A 0.91
(0.84-0.97)
0.93
(0.86–1)
19 /20
M
/A
M 0.94
(0.88-0.97)
1
(1-1)
24 /24
A 1
(1–1)
0.93
(0.86-0.97)
22 /24
Table 2: Classification cross-validation results measured in
a range between 0 and 1 in terms of ratio of correctly
classified patients for the AD vs MCI, Health vs AD and
MCI vs Healthy scenario, with all channels.
AD vs MCI Closed Eyes Open Eyes
AD MCI AD MCI
Hi
g
h Band 0.42 0.46 0.22 0.52
Low Band 0.92 1.00 0.93 1.00
Healthy vs AD Closed Eyes Open Eyes
Health AD Health AD
High Band 0.20 0.45 0.00 0.57
Low Band 1.00 0.95 0.90 1.00
MCI vs Health
y
Closed E
y
es O
p
en E
y
es
MCI Health MCI Health
High Band 0.70 0.00 0.76 0.00
Low Band 0.95 0.90 0.86 0.76
Table 3: Ratio of patients classified correctly in the three
binary classification scenarios, with selected channels.
AD vs MCI Closed Eyes Open Eyes
AD MCI AD MCI
Hi
g
h Band 0.04 0.58 0.07 0.52
Low Band 0.29 0.96 0.44 1.00
Health
y
vs AD Closed E
y
es O
p
en E
y
es
Health AD Health AD
High Band 0.50 0.40 0.00 0.48
Low Band 0.65 0.80 0.71 0.95
MCI vs Health
y
Closed E
y
es O
p
en E
y
es
MCI Health MCI Health
Hi
g
h Band 0.65 0.15 0.76 0.00
Low Band 0.90 0.00 1.00 0.00
More detailed results are presented in Table 2 and
Table 3, as regards the different schemes considered.
More specifically, these results show percentages of
correctly classified subjects per class and correspond
to the three binary classification models (based on the
2-stage classifier) and the schemes with 17 vs. 10
channels, open vs closed eyes, and high vs low-
frequency bands.
Figure 2: Classification results (correctly patients classified
ratio) with low / high band filter, with open / closed eyes
and all channels for top) AD vs MCI patients. mid) Healthy
vs AD patients, bottom) MCI vs Healthy patients.
The case with ten selected channels (in the
central-temporal zone) resulted in inferior results,
suggesting that the combined information from all
channels was useful. The case of Healthy vs. AD Low
Band Open Eyes and closed eyes is the only open-eye
case where classification results are quite high.
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
302

Figure 3: Classification results (correctly patients classified
ratio) with low/high band filter, with open/closed eyes and
all channels for top) AD vs. MCI patients. mid) Healthy vs.
AD patients, bottom) MCI vs. Healthy patients.
As illustrated in Figure 2, the scheme with the low
band filter and closed eyes works better for every case
in our dataset. In Healthy vs. AD and MCI vs. Healthy
examples, the algorithm returned in total the optimal
accuracy using the low band filter. Figure 3 presents
the correctly classified patients, when selected
channels are used, and performance is overall lower.
Overall, when considering together the results of
low-frequency closed and open eyes, only one AD
and one healthy subject are wrongly classified in both
open and closed eye cases, while one healthy subject
is wrongly classified vs. AD and vs. MCI in open
eyes. All other failures are basically in open eyes,
which is probably a more challenging case and lies in
the inconclusive area, having around 50% of patients’
segments in each class. The combination of both open
and closed eyes in a classification scheme might lead
to interesting results, and even manage to classify into
more subgroups.
Table 4: Summarised performance metrics for the 3-class
classification model. Precision and recall are calculated in
terms of percentages of correctly and falsely classified
segments per subject. Values correspond to median and 1
st
-
3d quartiles. The correctly classified patients are depicted
in the #C column.
Precision Recall #C
Healthy 0.96
0.86-1.0
0.85
0.72-0.96
16/20
MCI 0.75
0.69-0.87
0.95-
0.84-1.00
17/20
AD 1.00
0.99-1.00
0.95
0.90-0.98
19/20
In the case of the three-class classification
problem, Table 4 presents similar performance
metrics for the 3-class model in the testing set,
including segments from two subjects in each run.
Metrics include Median (1
st
-3
rd
quartile) for precision
and recall. Based on recall >0.6 in each run, the last
column shows the correctly classified subjects per
class. Results are slightly poorer in this case than the
binary models as presented in Table 1, especially
regarding the Healthy class. The 3-class model may
require more data for training.
Finally, an important issue that would need to be
addressed is that of exploring feature importance.
KPCA is not directly leading to insights about the
features that lead to best classification, and the
mechanisms behind that, and more sophisticated
methods would be required to illustrate results in
terms of interpretability.
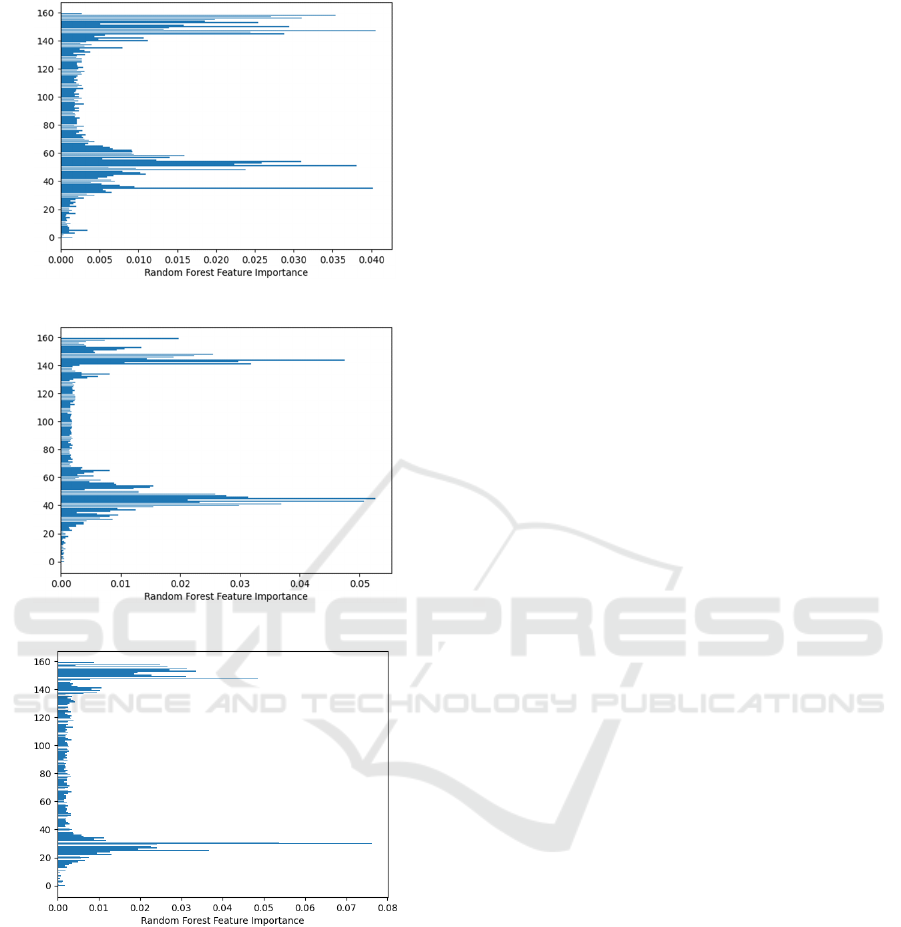
Nevertheless, Figure 4 provides feature
importance, as provided by the RF model, based on
the Gini importance, to illustrate the contribution of
multiple components of the KPCA transform, and
how these differ per classification problem. This
could potentially help in optimising the features
eventually selected in each classification model.
4 CONCLUSIONS
The presented method is based on KPCA for
dimensionality reduction of multichannel segments.
This method has been used before with EEG analysis
(Ye et al, 2018). Considering the classification
results, low band filter returns better accuracy for
both training and test set. Furthermore, the algorithm
works best with AD or MCI vs Healthy patients rather
than AD vs MCI. Compared to the results performed
in the comparative study of (Lehmann et al, 2007), a
rather higher accuracy is achieved. This is probably
because MCI and AD signals share some similarities,
Exploring Classification in Open and Closed Eyes EEG Data for People with Cognitive Disorders
303
(a)
(b)
(c)
Figure 4: Feature importance from the Random Forest
model classification results with low band filter, closed
eyes and all channels, for a) Healthy-MCI model, b) for
MCI-AD model, and c) Healthy-AD model. Y-axis
corresponds to 1-160 KPCA components used as features.
and the algorithm faces difficulty to correctly identify
patient data as one of those classes.
While the low frequency closed eyes scheme
seems to produce a better result than open eyes, it is a
matter of further research whether information from
both states would result in more stable and safe
results. Certainly, a larger training dataset and a more
comprehensive evaluation would improve the
credibility of the results. A more thorough finetuning
of the various parameters would also be of value and
would potentially lead to a more optimized outcome.
Furthermore, adding an explainability layer
would help better understand and trust the approach.
Finally, it would be relevant to address the problem
in a continuous space rather than a classification
problem and recognize the problem’s complexity
addressing the different subtypes of the MCI/AD
conditions.
REFERENCES
Amezquita-Sanchez, J. P., Mammone, N., Morabito, F. C.,
Marino, S., & Adeli, H. (2019). A novel methodology
for automated differential diagnosis of mild cognitive
impairment and the Alzheimer’s disease using EEG
signals. Journal of Neuroscience Methods, 322, 88–95.
https://doi.org/10.1016/j.jneumeth.2019.04.013
Awad, M., & Khanna, R. (2015). Support Vector Machines
for Classification. In Efficient Learning Machines (pp.
39–66). Apress. https://doi.org/10.1007/978-1-4302-
5990-9_3
Buegler M, Harms R, Balasa M, Meier IB, Exarchos T, Rai
L, Boyle R, Tort A, Kozori M, Lazarou E, Rampini M,
Cavaliere C, Vlamos P, Tsolaki M, Babiloni C,
Soricelli A, Frisoni G, Sanchez-Valle R, Whelan R,
Merlo-Pich E, Tarnanas I. 2020 Digital biomarker-
based individualized prognosis for people at risk of
Dementia. Alzheimers Dement (Amst). Aug
19;12(1):e12073. doi: 10.1002/dad2.12073. PMID:
32832589; PMCID: PMC7437401.
Dallora, A. L., Eivazzadeh, S., Mendes, E., Berglund, J., &
Anderberg, P. (2017). Machine learning and
microsimulation techniques on the prognosis of
Dementia: A systematic literature review. PLOS ONE,
12(6), e0179804. https://doi.org/10.1371/
journal.pone.0179804
Durongbhan, P., Zhao, Y., Chen, L., Zis, P., De Marco, M.,
Unwin, Z. C., Venneri, A., He, X., Li, S., Zhao, Y.,
Blackburn, D. J., & Sarrigiannis, P. G. (2019). A
Dementia Classification Framework Using Frequency
and Time-Frequency Features Based on EEG Signals.
IEEE Transactions on Neural Systems and
Rehabilitation Engineering, 27(5), 826–835. https://
doi.org/10.1109/TNSRE.2019.2909100
Kim, D., & Kim, K. (2018). Detection of Early Stage
Alzheimer’s Disease using EEG Relative Power with
Deep Neural Network. 2018 40th Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society (EMBC), 352–355. https://doi.org/
10.1109/EMBC.2018.8512231
Kulkarni, N., & Bairagi, V. K. (2014). Diagnosis of
Alzheimer Disease using EEG Signals. International
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
304

Journal of Engineering Research & Technology
(IJERT), 3(4).
Kumar, J. S., & Bhuvaneswari, P. (2012). Analysis of
Electroencephalography (EEG) Signals and Its
Categorization–A Study. Procedia Engineering, 38,
2525–2536. https://doi.org/10.1016/
j.proeng.2012.06.298
Lehmann C, Koenig T, Jelic V, Prichep L, et al (2007),
Application and comparison of classification
algorithms for recognition of Alzheimer’s disease in
electrical brain activity (EEG), Journal of Neuroscience
Methods, 161(2), 342-350,ISSN 0165-0270, https://
doi.org/10.1016/j.jneumeth.2006.10.023.
Raz, L., Knoefel, J., & Bhaskar, K. (2016). The
neuropathology and cerebrovascular mechanisms of
Dementia. Journal of Cerebral Blood Flow &
Metabolism, 36(1), 172–186. https://doi.org/10.1038/
jcbfm.2015.164
van der Maaten L., Hinton G. (2008). Visualizing data
using t-SNE. J. Mach. Learn. Res. 9, 2579–2605Wang,
Q. (2012). Kernel Principal Component Analysis and
its Applications in Face Recognition and Active Shape
Models. http://arxiv.org/abs/1207.3538
Ye B, Qiu T, Bai X, Liu P (2018). Research on Recognition
Method of Driving Fatigue State Based on Sample
Entropy and Kernel Principal Component Analysis.
Entropy (Basel). ;20(9):701. doi: 10.3390/e20090701.
PMID: 33265790; PMCID: PMC7513215.
Exploring Classification in Open and Closed Eyes EEG Data for People with Cognitive Disorders
305
