
Interpretable Disease Name Estimation based on Learned Models using
Semantic Representation Learning of Medical Terms
Ikuo Keshi
1,2
, Ryota Daimon
2
and Atsushi Hayashi
3
1
AI & IoT Center, Fukui University of Technology, 3-6-1, Gakuen, Fukui, Fukui, Japan
2
Electrical, Electronic and Computer Engineering Course, Department of Applied Science and Engineering,
Fukui University of Technology, 3-6-1, Gakuen, Fukui, Fukui, Japan
3
Department of Ophthalmology, University of Toyama, 2630 Sugitani, Toyama, Toyama, Japan
Keywords:
Interpretable Machine Learning, Semantic Representation Learning, Computer Assisted Coding, Discharge
Summary, Word Semantic Vector Dictionary, Disease Thesaurus.
Abstract:
This paper describes a method for constructing a learned model for estimating disease names using semantic
representation learning for medical terms and an interpretable disease-name estimation method based on the
model. Experiments were conducted using old and new electronic medical records from Toyama University
Hospital, where the data distribution of disease names differs significantly. The F1-score of the disease name
estimation was improved by about 10 points compared with the conventional method using a general word
semantic vector dictionary with a faster linear SVM. In terms of the interpretability of the estimation, it was
confirmed that 70% of the disease names could provide higher-level concepts as the basis for disease name
estimation. As a result of the experiments, we confirmed that both interpretability and accuracy for disease
name estimation are possible to some extent.
1 INTRODUCTION
Because interpreting learning results with neural net-
works is complex and a large amount of training data
is required, there have been challenges in applying
neural networks to disease name estimation for dis-
charge summaries, which often have a small num-
ber of cases of disease names. Disease name esti-
mation is the automatic assignment of ICD10 codes
from the standard disease name master to discharge
summaries.
In the United States, tools to assist medical infor-
mation managers with ICD10 coding tasks have al-
ready been commercialized and are becoming more
widely used. It is believed that ICD10 codes are
derived by applying natural language processing to
medical documents, but the algorithm has not been
disclosed. In addition, no similar support tool us-
ing natural language processing has been developed
in Japan (Tsujioka et al., 2022).
We previously proposed a semantic representation
learning method that improves the accuracy of docu-
ment classification and the interpretability of learn-
ing results even in the absence of sufficient training
data by representing finite numbers of hidden nodes
in a neural network with feature words representing
meanings. We introduced a word semantic vector dic-
tionary as an initial value for weights between words
and hidden nodes (Keshi et al., 2017; Keshi et al.,
2018). The dictionary is a general-purpose dictio-
nary describing the relationship between 264 feature
words selected on the basis of an encyclopedia and
20,000 core words (Keshi et al., 1996). The 264 fea-
ture words correspond to 264 concept classifications
in the encyclopedia. Therefore, it is possible to ob-
tain a distributed representation that people can inter-
pret. We also proposed a method for constructing a
Japanese version of CAC (Computer Assisted Cod-
ing: an ICD10 coding support tool for medical in-
formation managers) using the general-purpose word
semantic vector dictionary (Tsujioka et al., 2022).
This study aimed to adapt the word semantic vec-
tor dictionary to estimate disease names in the medi-
cal field and improve the performance of interpretable
disease-name estimation. In this paper, we show
that 264 disease-name feature words selected from
a disease name thesaurus improve the accuracy of
disease name estimation with the CAC construction
method by about 10 points relative to the F1-score
with a faster linear SVM, compared with the general-
Keshi, I., Daimon, R. and Hayashi, A.
Interpretable Disease Name Estimation based on Learned Models using Semantic Representation Learning of Medical Terms.
DOI: 10.5220/0011548900003335
In Proceedings of the 14th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2022) - Volume 1: KDIR, pages 265-272
ISBN: 978-989-758-614-9; ISSN: 2184-3228
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
265

purpose word semantic vector dictionary. In addition,
the results of an interpretability evaluation using the
visual statistics software StatFlex show that seman-
tic representation learning of progress summaries can
present higher-level concepts of disease names as a
basis for disease name estimation.
2 RELATED WORK
In 2013, Mikolov et al. presented word2vec, which
uses neural networks to learn contextual information
from text (Mikolov et al., 2013a; Mikolov et al.,
2013b; Mikolov et al., 2013c). They reported that
vectors with similar weights (distributed representa-
tions) could be constructed when a neural network
learns words with similar meanings. The problem
with the distributed representations acquired by neu-
ral networks is that it is difficult to know which mean-
ing each dimension corresponds to, and the meaning
of each dimension changes in each learning domain.
In addition, since a large amount of text data is re-
quired for training, accuracy cannot be achieved with
a small amount of data in each domain.
As for studies on assigning meaning to each axis
of a distributed representation of words, the inter-
pretability of word2vec, which starts word learning
from random initial values, has been studied with the
non-negative online learning Skip-gram model (Luo
et al., 2015) and the sparse constrained online learn-
ing SparseCBOW model (Sun et al., 2016).
In the study of disease coding, a previous
study (Suzuki Takahiro, 2019) attempted to automati-
cally determine the DPC (Diagnosis Procedure Com-
bination) from cases using a vector space model for
more than 20 cases of DPC in a discharge summary.
The advantages and differences of the proposed
method are shown below.
• When learning distributed representations of
words in the Wikipedia corpus, related stud-
ies (Luo et al., 2015) (Sun et al., 2016) sorted
about 500,000 words by dimension, and a per-
son had to analogize the meaning of each dimen-
sion from the top words. Also, the meaning of
the dimension changes depending on the research
domain. The proposed method is easy to use
because 264 disease-name feature words express
the meaning of each dimension. In addition, re-
lated studies evaluated only word similarity, and
the proposed method is also effective in extract-
ing document features, such as in disease name
estimation.
• In a related study (Suzuki Takahiro, 2019), it was
adequate to determine DPC codes from cases of
several hundred words, but it was challenging to
estimate disease names from chief complaints of a
few words. Since the proposed method expresses
the meaning of a word or a case with the same
264 feature words, it is effective for estimating the
name of a disease from a chief complaint.
3 PROPOSED METHOD
The feature of our proposed CAC (Tsujioka et al.,
2022) is that it uses support vector machines (SVMs)
after converting the progress summary of a discharge
summary into 264 feature-word vector values in a dis-
tributed representation using a neural network, which
is called semantic representation learning. A charac-
teristic of semantic representation learning is that it
can be quantified in a form easily understood by hu-
mans, making this method highly compatible with the
medical field, where accountability is a crucial issue.
This paper proposes a method for applying seman-
tic representation learning to specialized fields such
as medicine. Instead of using a word semantic vector
dictionary manually constructed from an encyclope-
dia as a seed vector for the neural network, a medical-
word semantic-vector dictionary automatically built
from a thesaurus of disease names is introduced. The
definition of interpretability in this paper is that peo-
ple can understand the meaning of a feature word with
a significant weight in the distributed representation
and that the feature word provides a basis for disease
names estimated by SVM.
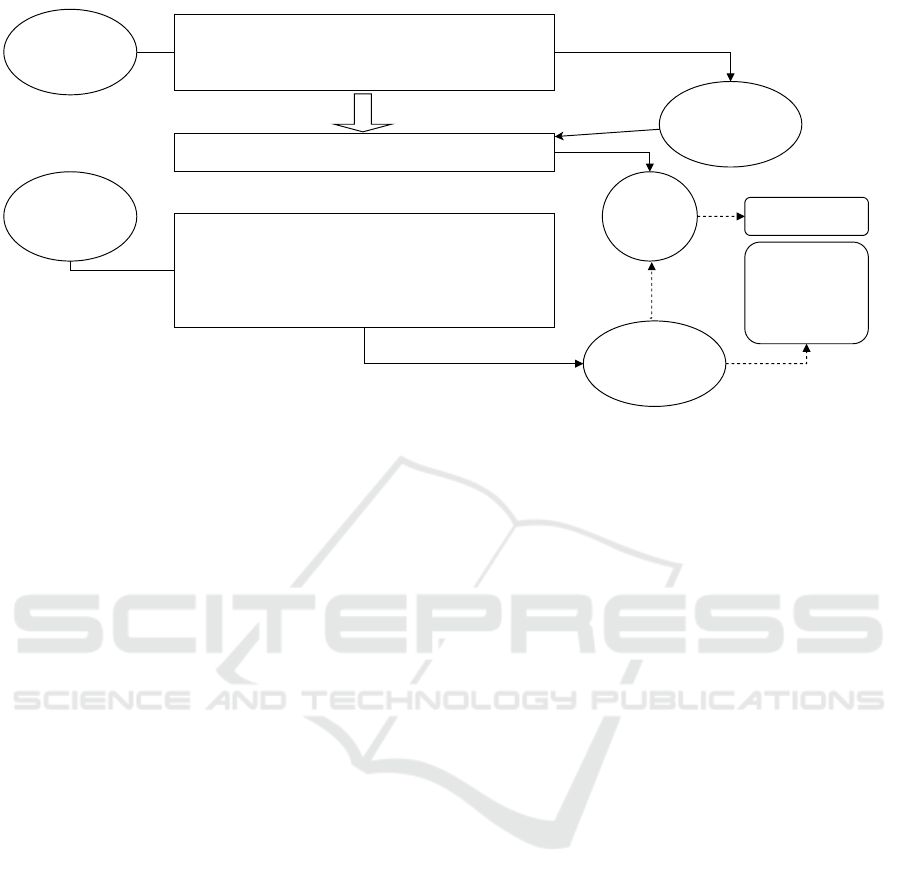
As shown in Fig. 1, the disease-name esti-
mation method performs (1) medical-word seman-
tic representation learning and (2) learned model
creation for estimating disease names on electronic
medical records for training. In addition, (3) inter-
pretable disease-name estimation is performed on the
electronic medical records for evaluation, and dis-
ease name codes are obtained by referring to the
learned disease-name estimation model. Interpretable
disease-name estimation can obtain disease names
(feature words) that are higher-level concepts of the
disease name codes by selecting feature words with
the highest weights from the weight vectors.
In this Section 3, the following subsections de-
scribe in turn (1) medical-word semantic representa-
tion learning, (2) learned model creation for estimat-
ing disease names, and (3) interpretable disease-name
estimation, which constitute the method for estimat-
ing interpretable disease names.
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
266
(1) Medical-word semantic representation learning
(A) Data processing of the target sentences
(B) Creation of a word semantic vector dictionary
(C) Representing the target sentences by N feature words and obtaining
their weight vectors.
(2) Creating a learned model for estimating disease names
(D) Machine learning
(3) Estimating interpretable disease names
(A) Data processing of the target sentences
(B) Creation of a word semantic vector dictionary
(C) Representing the target sentences by N feature words and obtaining
their weight vectors.
(E) Estimating the disease name code and obtaining the upper concept of
the disease name
Accumulated
electronic medical
records for training
Electronic medical
records for evaluation
Learned
disease-name
estimation
model
Weight vectors of the
electronic medical
records for evaluation
Weight vectors of all
electronic medical
records for training
Reference
Disease name code
Disease names that
are higher level
concepts of the
above disease
name codes
Selecting feature words with
the highest weights
Figure 1: Interpretable disease-name estimation methods.
3.1 Medical-word Semantic
Representation Learning
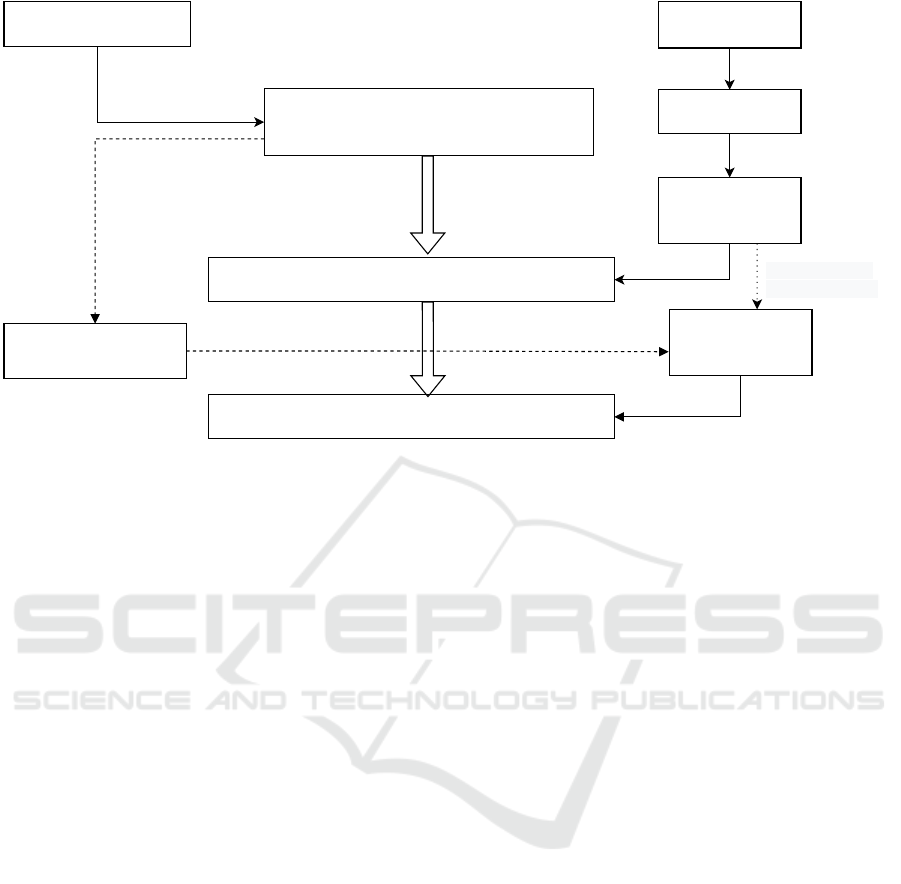
The semantic representation learning of medical
terms is shown in Fig. 2.
(A) Data processing of progress summaries,
(B) Creation of a medical-word semantic-vector dic-
tionary representing disease names as N feature
words,
(C) Representing target sentences as N feature words
and obtaining their weight vectors.
The details of these steps (A) through (C) are as
follows. First, (A) data processing of the progress
summaries is as follows.
(A-1) Prepare an electronic medical record with a
discharge summary that includes the diagnosed per-
son’s “sex,” “age,” “department name,” the diagnosed
disease name expressed by the disease code, and a
progress summary. The progress summary in the dis-
charge summary summarizes the patient’s chief com-
plaint, medical history, physical examination find-
ings, and medical treatment details during hospital-
ization.
(A-2) Use morphological analysis to obtain a word-
for-word segmentation of progress summaries.
Second, (B) creation of a word semantic vector
dictionary is as follows.
(B-1) Prepare a thesaurus that contains relationships,
synonyms, and disease names.
(B-2) Select N feature words from the thesaurus.
(B-3) For all the disease names registered in the the-
saurus, list the feature words corresponding to the
superordinate relations, synonyms, and the disease
names to obtain a word semantic vector dictionary.
Furthermore, (C) obtaining the weight vector of
the target text is as follows.
(C-1) Using the word semantic vector dictionary, rep-
resent all words appearing in the progress summary as
the feature words and assign a seed vector consisting
of N vector values. The step of assigning a seed vector
to all words includes the step of recursively expand-
ing all said words represented by said N feature words
using said word semantic vector dictionary (Faruqui
et al., 2015).
(C-2) Learn N types of vector values for each progress
summary, and obtain a weight vector for the progress
summary. The step of obtaining a weight vector of
the progress summary includes obtaining a paragraph
vector (Le and Mikolov, 2014) represented by N vec-
tor values of the progress summary for each electronic
medical record using said seed vector.
As explained above, this medical-word semantic
representation learning uses a word semantic vector
dictionary created from a thesaurus of disease names
to obtain a weight vector of progress summaries. This
weight vector is represented by these N types of vec-
tor values in a space stretched by N feature words of
higher-level concepts selected from the thesaurus of
disease names.
3.2 Creating Learned Model for
Estimating Disease Names
In this section, we use the medical-word semantic rep-
resentation learning described in Section 3.1 to obtain
the weight vector of the progress summaries of elec-
tronic medical records for training and perform ma-
chine learning on it to create a learned model for dis-
Interpretable Disease Name Estimation based on Learned Models using Semantic Representation Learning of Medical Terms
267
(A) Data processing of progress summaries
(B) Creation of a medical-word semantic-vector dictionary
(C) Representing the target sentences by N feature words
and obtaining their weight vectors
Electronic medical records
Progress summaries
Separating words
in Japanese with
spaces
Disease thesaurus
Select N feature
words
Creation of word
semantic vector
dictionary
Assigning an N-
dimensional seed
vector to every word
Word semantic
vector dictionary
Learning N-dimensional vector values
for each progress summary
All words in the progress
summary are represented
by feature words
Figure 2: Medical-word semantic representation learning method.
ease name estimation. The method is as follows (see
Fig. 1). The method for creating a learned model
for disease name estimation uses information, includ-
ing the weight vector of the progress summary of an
electronic medical record for training, obtained as an
explanatory variable. By using the information, in-
cluding the vector as an explanatory variable, and per-
forming machine learning, the weight vector of the
explanatory variable can be made to correspond to
the disease code of the diagnosed disease name in the
electronic medical record for training as an objective
variable. Here, the explanatory variables also include
“gender,” “age,” and “department name” in the elec-
tronic medical record. In creating a learned disease
model, step (D) for performing machine learning is
as follows.
(D-1) Select the top M diagnostic disease names from
the electronic medical records for evaluation.
(D-2) Input the explanatory variables of all the elec-
tronic medical records for training with the selected
M diagnostic disease names and their corresponding
objective variables into an SVM.
As described above, the method for creating a
disease-name learned model can obtain, as an objec-
tive variable, a disease name code for a diagnostic dis-
ease name corresponding to an explanation vector by
performing machine learning.
3.3 Estimating Interpretable Disease
Names
By referring to the learned disease-name model ob-
tained for the electronic medical records for training
in Section 3.2, the disease-name estimation method
estimates disease name codes for electronic medical
records for evaluation that have undergone the word
semantic representation learning described in Section
3.1, and it also obtains disease names (feature words)
that are higher-level concepts. Thus, the method for
estimating disease names is as follows.
• (Step 1) Perform steps (A) to (C) on the prepared
electronic medical record for evaluation to obtain
the weight vectors of the explanatory variables.
• (Step 2) Perform steps (A) to (C) on the prepared
electronic medical record for training to obtain the
weight vectors of the explanatory variables.
• (Step 3) Perform step (D) on the explanatory vari-
ables and their corresponding objective variables
of the electronic medical record for training to ob-
tain a learned model of the disease names.
• (Step 4) Perform step (E) on the explanatory vari-
ables of the electronic medical record for evalu-
ation to estimate the disease name codes by re-
ferring to the learned model of disease names in
the electronic medical record for training obtained
in (Step 3). The disease names (feature words),
higher-level concepts of the disease codes, are
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
268
also estimated and obtained from the weight vec-
tors.
The interpretable disease-name estimation
method can obtain the disease name of the feature
word, an upper concept of the disease name code
obtained from the disease-name learned model, by
executing Step 1 to Step 4.
4 EXPERIMENTAL SETUP
4.1 Preparation of Electronic Medical
Records
In these experiments, the electronic medical records
for training and evaluation were discharge summaries
from 2004-2019 from the University of Toyama Hos-
pital. The electronic medical record for training
was in the form of the old electronic medical record
(NeoChart) for 2004-2014, with 94,083 records and
3,204 total disease names. The electronic medical
record for evaluation was in the form of the new
electronic medical record (EGMAIN-GX) for 2015-
2019, with 61,772 records and 2,849 total disease
names. This new record for evaluation is abbreviated
as EGMAIN-GX and the old record for training as
NeoChart. In addition, the following records and oth-
ers were deleted as conditions for data cleansing.
• Records with missing values.
• Fields not used as explanatory variables.
• Rare disease names that represented less than
0.02% of the total number of records.
• Records with progress summaries of less than 50
characters.
Table 1 shows the number of records in NeoChart
with the top 20 (M=20) diagnosis disease codes in
EGMAIN-GX. Although there is a significant differ-
ence in the distribution between the two, it was found
that the top 20 disease codes in EGMAIN-GX were
also present in NeoChart for more than 70 or more
cases. Also, in the case of M=20, there are more than
10,000 cases in both electronic medical records, en-
suring an adequate number of records for SVM train-
ing and evaluation.
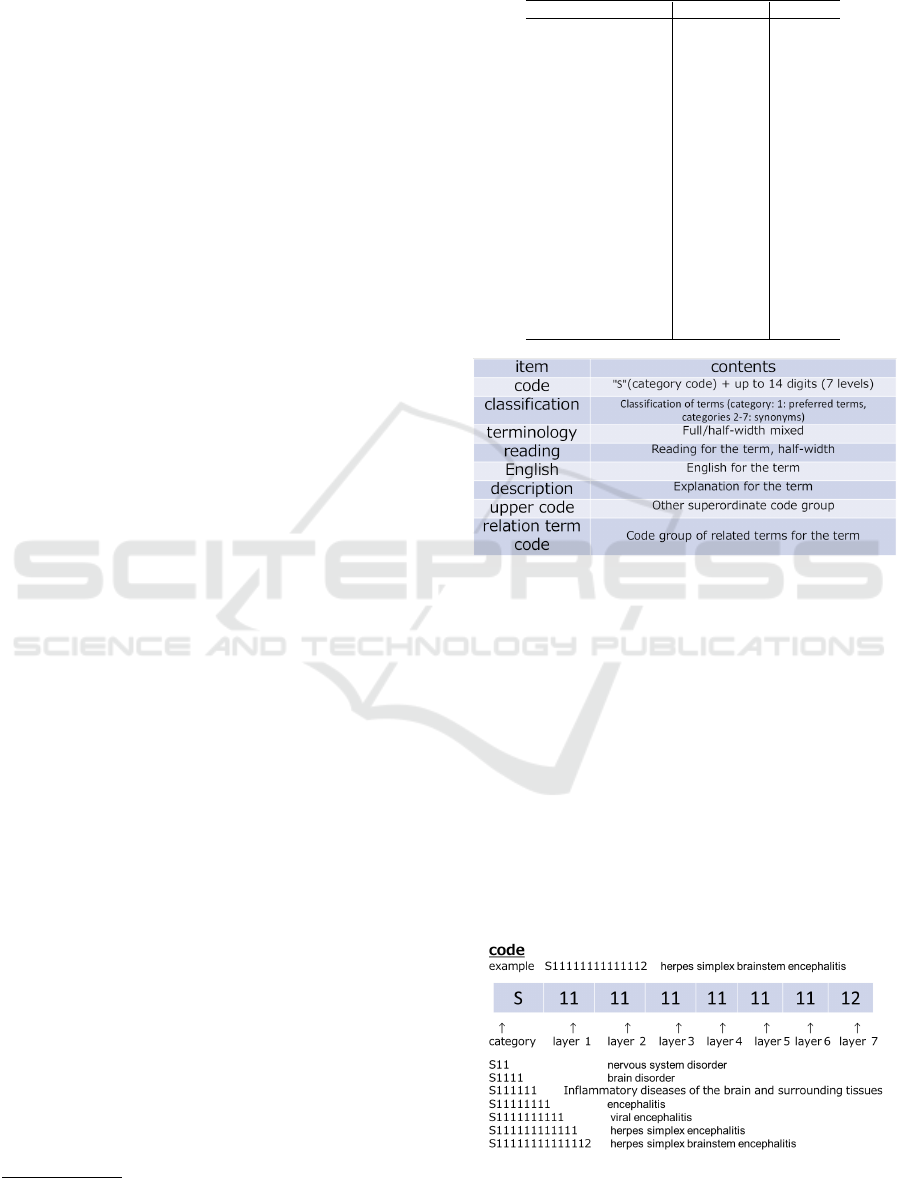
4.2 Preparation of Disease Thesaurus
For the disease thesaurus, we used the T-dictionary
1
,
which is structured as shown in Fig. 3. The item
1
https://www.tdic.co.jp/products/tdic
Table 1: Number of cases in NeoChart corresponding to top
20 diagnosis disease codes in EGMAIN-GX.
Diagnosis disease code EGMAIN-GX NeoChart
C34.1 1127 210
H25.1 929 123
C61 912 2216
C34.3 893 158
C22.0 864 1501
I20.8 698 75
I35.0 690 70
I50.0 545 166
C16.2 536 231
I67.1 515 387
C25.0 503 111
C15.1 483 253
I48 483 253
C34.9 468 1579
P03.4 432 399
C56 393 1276
M48.06 373 845
H35.3 368 1060
H33.0 361 625
C20 357 343
Figure 3: Item structure in disease thesaurus (T-dictionary).
“code” in the top row is “S (category code) + num-
ber up to 14 digits (7 levels),” as shown in the ex-
ample listed in Fig. 4. The item “category” in the
second row is the classification of terms (category 1:
preferred terms, categories 2 to 7: synonyms).
4.3 Creation of Medical-word
Semantic-vector Dictionaries
For constructing word semantic vector dictionaries
from the T-dictionary, 264 disease names (N=264)
were first selected as feature words to compare them
with our conventional method using an encyclopedia.
Figure 4: Structure of disease codes in disease thesaurus
(T-dictionary).
Interpretable Disease Name Estimation based on Learned Models using Semantic Representation Learning of Medical Terms
269

nervous system
disorder
nervous system
disorder
encephalopathy・
neurological
disorder
nervous system
disorder
neurological
disease
nervous system
disorder
nervous system
disease
nervous system
disorder
encephalopathy nervous system
disorder
encephalopathy
cerebral
disease
nervous system
disorder
encephalopathy
brain
disease
nervous system
disorder
encephalopathy
Encephalopathy is the ICD10 standard form of brain disease belonging to the second tier.
Disease feature words
nervous system disorder
digestive tract disorder
cardiovascular disorder
encephalopathy
Disease names Related disease feature words
Figure 5: Example of medical-word semantic-vector dictio-
nary.
The 264 disease feature words extracted from the T-
dictionary were selected from preferred terms of five
letters or less in seven levels; the higher the concept
of the disease name in the T-dictionary, the more re-
lated words (synonyms and subordinate words) were
registered. Similarly, to test the effect of increasing
the number of disease feature words on the accuracy
of disease name estimation, all preferred terms with
five or fewer letters in the first through sixth levels
of the T-dictionary were extracted as 458 disease fea-
ture words. In these experiments, disease names in
the T-dictionary were used to create a medical-word
semantic-vector dictionary, as shown in Fig. 5. In
case (N=264), eight feature words were duplicated
by converting to the standard ICD10 format, and the
number of disease names decreased from 36,768 to
31,033 words.
Figure 6: Experimental flow of disease name estimation for
two electronic medical records with different data distribu-
tions.
Table 2: Evaluation results: macro average F1-score for es-
timating disease names.
Feature words Linear SVM Linear SVM (SMOTE)
264 random weights (doc2vec) 38.4 35.7
264 concept classification 59.8 62.6
264 disease names 71.2 72.4
458 disease names 71.5 70.6
5 EVALUATION RESULTS
5.1 Disease Name Estimation
As shown in the flow diagram in Fig. 6, the semantic
representation learning was performed on EGMAIN-
GX and NeoChart, from which the weight vectors of
all electronic medical records (progress summaries)
of both were obtained. Next, training data (11,839
cases) were created for the top 20 disease names in
EGMAIN-GX from the 73,150 cases in NeoChart af-
ter data cleansing. The weight vectors of all electronic
medical records (progress summaries) of both were
obtained, and machine learning (left column of Fig.
6) was performed to obtain a disease-name learned
model (bottom center of Fig. 6).
Next, as shown in the flow diagram in Fig. 6, eval-
uation data were created for the top 20 disease names
(11,931 cases) of the 48,911 cases in EGMAIN-GX.
The disease name estimation was performed with ref-
erence to the disease-name learned model (right col-
umn of Fig. 6), and the estimation accuracy (macro
average F1-score) was evaluated. On the basis of the
progress summary’s weight vector, explanatory vari-
ables (age, gender, and department) were added. The
accuracy (F1-score) of the disease name estimation
was evaluated using the linear SVM.
Table 2 summarizes the results (macro average
F1-score). First shown are the results of doc2vec,
which is Gensim’s library
2
that implements para-
graph vectors (Le and Mikolov, 2014) and learns
words and paragraphs from random initial weights.
The macro average F1-score of NeoChart’s disease-
name estimation was over 90, whereas the F1-score
of EGMAIN-GX’s disease-name estimation using
NeoChat’s learned model was much lower at 38.4.
The hyperparameters of doc2vec were 264 dimen-
sions, word order was not considered, and the num-
ber of epochs was set to 20. A grid search for linear
SVM was performed on the basis of the evaluation re-
sults of the development set of NeoChart, and the cost
parameter C was set to 0.02. Next, Table 2 shows the
results of encyclopedia concept classifications used as
feature words. This is our conventional method. Fi-
nally, the table shows the results when 264 and 458
2
https://radimrehurek.com/gensim/models/doc2vec.html
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
270
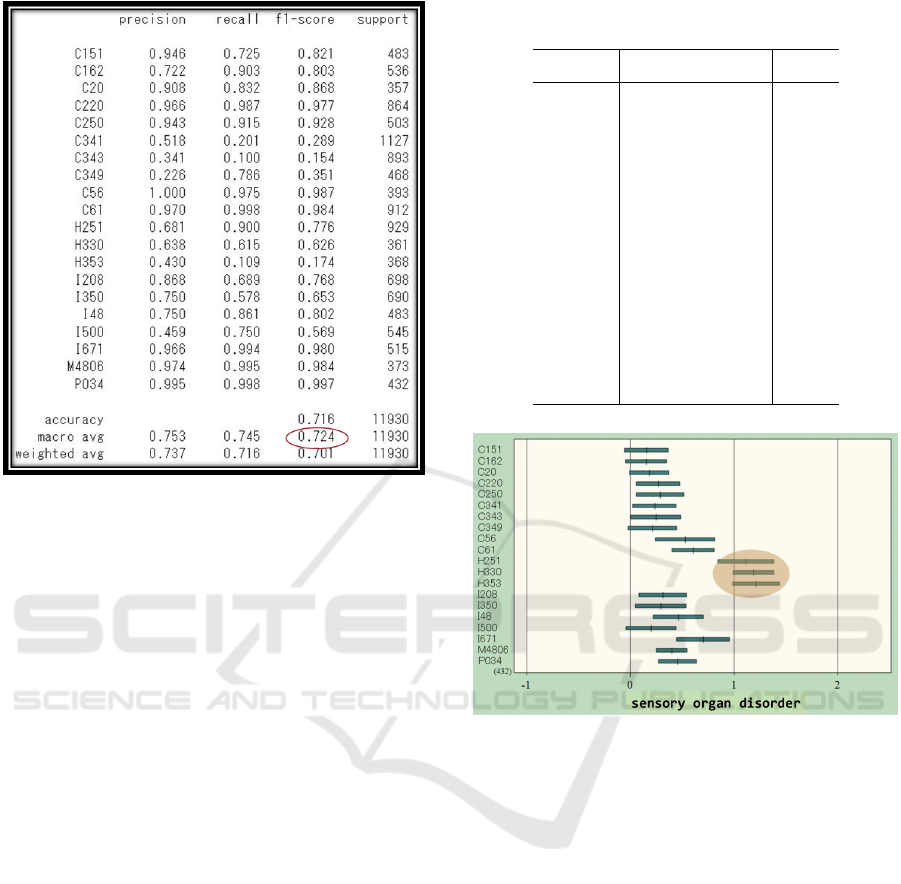
Figure 7: Evaluation results for disease name estimation us-
ing linear SVM with SMOTE.
disease names were used as feature words.
Table 2 shows that the F1-score for disease name
estimation when 264 feature words were selected
from the disease thesaurus was about 10 points higher
than that of the conventional method. The re-
sults were almost identical when 458 and 264 fea-
ture words were selected from the disease thesaurus.
When 264 feature words were selected, the training
set was expanded to the same number of pieces of
data for each disease name by SMOTE (Lema
ˆ
ıtre
et al., 2017), and the disease estimation model was
constructed by linear SVM, the F1-score of the eval-
uation set was 72.4, indicating that it was the best
macro average F1-score. The hyperparameters, in this
case, were 5 epochs of word learning and 20 epochs
of paragraph vector learning for semantic representa-
tion learning, and the C parameter was 0.03 for SVM.
SMOTE is a method of oversampling data to align
the number of pieces of data in each classification
class when the number in each class is unbalanced.
Fig. 7 shows the evaluation results of the F1-score
for disease name estimation using linear SVM with
SMOTE.
5.2 Interpretability
We evaluated the interpretability of semantic repre-
sentation learning using the 264 disease-name fea-
ture words. Regarding the top 20 disease codes in
EGMAIN-GX, Table 3 shows the feature words with
the highest weights among the 264-dimensional vec-
Table 3: Interpretability of disease name estimation and its
success or failure.
Diagnosis feature words Results
disease code with the highest weights
C15.1 digestive tract disorder correct
C16.2 digestive tract disorder correct
C20 digestive tract disorder correct
C22.0 hepatic disease correct
C25.0 digestive tract disorder correct
C34.1 cardiovascular disorder incorrect
C34.3 cardiovascular disorder incorrect
C34.9 cardiovascular disorder incorrect
C56 blood disease incorrect
C61 blood disease incorrect
H25.1 sensory organ disorder correct
H33.0 sensory organ disorder correct
H35.3 sensory organ disorder correct
I20.8 cardiovascular disorder correct
I35.0 cardiovascular disorder correct
I48 cardiovascular disorder correct
I50.0 cardiovascular disorder correct
I67.1 cardiovascular disorder correct
M48.06 cardiovascular disorder incorrect
P03.4 neonatal disorder correct
Figure 8: Distribution of weights by disease code for sen-
sory organ disorders.
tor values obtained by semantic representation learn-
ing of the progress summaries. That is, the six fea-
ture words with the highest weights were “sensory
organ disorder,” “neonatal disorder,” “digestive tract
disorder,” “cardiovascular disorder,” “hepatic disor-
der,” and “blood disorder.” The correct rate of inter-
pretability of the 20 disease codes was 70%.
Here, we visualized the distribution of the weights
of progress summaries by disease code for the fea-
ture words. Fig. 8, which visualizes the weight dis-
tribution of sensory organ disorders by disease code,
shows that the weight of the feature word “sensory or-
gan disorder” was particularly high for the presump-
tive disease codes H251 “senile nuclear cataract,”
H330 “retinal detachment, retinal tear,” and H353
“degeneration of macula and posterior pole.” The re-
sults of the interpretability evaluation showed that se-
mantic representation learning of progress summaries
could provide higher-level concepts of disease names
as a basis for disease name estimation.
Interpretable Disease Name Estimation based on Learned Models using Semantic Representation Learning of Medical Terms
271

6 CONCLUSION
In this study, we introduced a disease thesaurus as a
seed vector for semantic representation learning us-
ing a CAC construction method. We showed that
by selecting 264 disease-name feature words, the F1-
score of disease name estimation was 72.4, which
is about 10 points more accurate than the general-
purpose word semantic vector dictionary with a faster
linear SVM. We also showed that semantic represen-
tation learning of progress summaries in electronic
medical records could provide higher-level concepts
of disease names as a basis for disease name estima-
tion. The accuracy was 70%. The reason for the fail-
ure in estimating the higher-level concepts of the pre-
sumed disease names was that the higher-level con-
cepts of those disease names were not included in
the feature words due to the setting of disease names
with five or fewer letters in the selection of feature
words. Adding these correct disease names to the fea-
ture words could significantly improve accuracy.
Comparative experiments on disease name es-
timation using doc2vec showed that although dis-
tributed representation learning can be adapted to a
given corpus, the accuracy of disease name estima-
tion is significantly degraded by learned models with
significantly different data distributions. Although
the proposed method was able to solve this prob-
lem, the F1-score needs to be further improved for
practical use. In the future, we plan to integrate our
method with a Bert/transfer model (Yoshimasa Kawa-
zoe, 2021) learned from a large number of Japanese
medical texts to improve the accuracy of estimating
interpretable disease names to a practical level.
ACKNOWLEDGEMENTS
This work was supported by JSPS KAKENHI Grant
Number 20K11833. This study was approved by the
Ethical Review Committee of the Fukui University of
Technology and the Toyama University Hospital.
REFERENCES
Faruqui, M., Dodge, J., Jauhar, S. K., Dyer, C., Hovy, E.,
and Smith, N. A. (2015). Retrofitting word vectors
to semantic lexicons. In Proc. of NAACL-HLT, pages
1606–1615.
Keshi, I., Ikeuchi, H., and Kuromusha, K. (1996). Associa-
tive image retrieval using knowledge in encyclopedia
text. Systems and Computers in Japan, 27(12):53–62.
Keshi, I., Suzuki, Y., Yoshino, K., and Nakamura, S.
(2017). Semantically readable distributed represen-
tation learning for social media mining. In Proc. of
IEEE/WIC/ACM International Conference on Web In-
telligence (WI), pages 716–722.
Keshi, I., Suzuki, Y., Yoshino, K., and Nakamura, S.
(2018). Semantically readable distributed represen-
tation learning and its expandability using a word se-
mantic vector dictionary. IEICE TRANSACTIONS on
Information and Systems, E101-D(4):1066–1078.
Le, Q. V. and Mikolov, T. (2014). Distributed representa-
tions of sentences and documents. In Proc. of ICML,
pages 1188–1196.
Lema
ˆ
ıtre, G., Nogueira, F., and Aridas, C. K. (2017).
Imbalanced-learn: A python toolbox to tackle the
curse of imbalanced datasets in machine learning.
Journal of Machine Learning Research, 18(17):1–5.
Luo, H., Liu, Z., Luan, H., and Sun, M. (2015). Online
Learning of Interpretable Word Embeddings. In Proc.
of EMNLP, pages 1687–1692.
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013a).
Efficient estimation of word representations in vector
space. CoRR, abs/1301.3781.
Mikolov, T., Sutskever, I., Chen, K., Corrado, G., and
Dean, J. (2013b). Distributed representations of
words and phrases and their compositionality. CoRR,
abs/1310.4546.
Mikolov, T., Yih, W., and Zweig, G. (2013c). Linguistic
regularities in continuous space word representations.
In Proc. of NAACL, pages 746–751.
Sun, F., Guo, J., Lan, Y., Xu, J., and Cheng, X. (2016).
Sparse Word Embeddings Using `
1
Regularized On-
line Learning. In Proc. of IJCAI, pages 2915–2921.
Suzuki Takahiro, Doi Shunsuke, C. K. K. T. S. K.-i. S. G.
N. R. H. Y. H. M. M. Y. M. T. Y. H. K. E. (2019).
Development of discharge summary audit support ap-
plication by text mining. In Proc. of Japan Assciation
for Medical Informatics, volume 39, pages 667–668.
Tsujioka, K., Keshi, I., Nakagawa, H., and Hayashi, A.
(2022). Research on a method for constructing a
Japanese version of computer assisted coding us-
ing natural language processing. Health Information
Management, 34(1):56–64.
Yoshimasa Kawazoe, Daisaku Shibata, E. S. E. A. K. O.
(2021). A clinical specific bert developed using a huge
japanese clinical text corpus. PLoS One, 16(11)(9).
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
272
