
Identification of MLST8, a Component of MTOR Complex 1 in
Eriocheir Sinensis: CDNA Cloning and Its Expression Respond to
Starvation
Zhihuan Tian
1
and Chuanzhen Jiao
*2
Henry Fok School of Biology and Agriculture, Shaoguan University, Shaoguan, China
*
Corresponding author
Keywords: Crustacean, MLST8, MTOR, Starvation.
Abstract: The mechanistic target of rapamycin (mTOR) signaling pathway is conserved among organisms from single-
celled yeasts to complex multicellular ones such as human. Here we report the cDNA sequences and
expression pattern of one gene encoding a component of mTOR complex 1 from crustacean Eriocheir sinensis,
EsmLST8 (mammalian lethal with Sec13 protein 8). The deduced EsmLST8 from the cDNA sequence
includes 317 amino acids (aa), which contains a WD40 superfamily region with WD40 repeats domains,
forming a circularized beta-propeller structure. Expression analysis with qRT-PCR showed that in juvenile E.
sinensis, the top three tissues with high mRNA levels for EsmLST8 are Y organ, stomach and hepatopancreas;
the tissue with the lowest expression of mLST8 is eyestalk. After the animals’ food deprivation, the expression
of EsmLST8 was significantly induced at 14d in claw muscles. These results provide basic information on the
functions of mTOR signaling pathway in regulation of growth and nutritional metabolism in crustaceans.
1 INTRODUCTION
The mechanistic target of rapamycin (mTOR) is an
conservative serine/threonine kinase of PI3K-related
kinase (PIKK) family from single-celled yeast to
multiple-celled complex animals such as human,
which form two functionally distinct protein
complexes of mTOR complexes: Complexes 1
(mTORC1) and 2 (mTORC2) (Yang et al., 2013). The
mTORC1 responds to extracellular stimulus such as
growth factors, energy, oxygen, amino acids and
mechanical stimulus, and is involved in regulating
protein, lipid, nucleotide, and glucose metabolism in
mammals (Chen and Long, 2018). It plays a central
role in controlling the balance between anabolism and
catabolism of mammals in response to environmental
conditions (Saxton and Sabatini, 2017).
The mLST8 (mammalian lethal with Sec13
protein 8), together with mTOR and raptor (regulatory
protein associated with mTOR), are three core
components of the mTORC1 (Yonezawa et al., 2004).
The mLST8 plays critical roles in mTOR kinase
1
https://orcid.org/0000-0001-9249-5036
2
https://orcid.org/0000-0002-8946-9619
activity by associating with its catalytic domain and
stabilizing the kinase activation loop (Yang et al.,
2013).
Crustaceans undergo periodic molting during
their growth. The molting is controlled by steroid
hormone ecdysteroids secreted by YO (Y-organ,
crustacean molting gland) and molt-inhibiting
hormone (MIH) neuropeptides secreted by X-
organ/sinus gland complex in eyestalk (Mykles, 2011;
Webster et al., 2012). Inhibited by MIH neuropeptides,
the ecdysteroids are in low level in hemolymph at the
stages of inter-molt and post-molt. The mTOR
signaling pathway genes’ transcripts were well
present in YO by transcriptome analyses (Das et al.,
2016); and the biosynthesis of ecdysteroids in YO
requires mTOR-dependent protein synthesis in early
pre-molt stage in crustacean (Abuhagr et al., 2016;
Das et al., 2018). In black land crab, the mTOR
signaling genes were up-regulated to activate YO and
sustain ecdysteroidogenesis in mid- and late pre-molt
stages (Abuhagr et al., 2014; Shyamal et al., 2018).
Investigating the different components of mTOR
Tian, Z. and Jiao, C.
Identification of MLST8, a Component of MTOR Complex 1 in Eriocheir Sinensis: CDNA Cloning and Its Expression Respond to Starvation.
DOI: 10.5220/0011594400003430
In Proceedings of the 8th International Conference on Agricultural and Biological Sciences (ABS 2022), pages 11-18
ISBN: 978-989-758-607-1; ISSN: 2795-5893
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
11

pathway is the necessary step to study the function of
this pathway, while its most components in crustacean
Eriocheir sinensis have not been intensively studied.
In this study we cloned the cDNA sequence encoding
a mTOR pathway component mLST8 (EsmLST8) in
E. sinensis, and their transcriptional expression were
investigated when animals under conditions of
starvation.
2 MATERIALS AND METHODS
2.1 Animal Sampling
One-year-old juvenile crabs, weighing 10.95 ± 2.25 g,
obtained from Gucheng fisheries farm (Jiangsu
Province, China) in May, 2019. The crabs were
maintained in tanks containing freshwater
approximately 2 cm in depth with the nature
photoperiod and the temperature of 25 ± 2°C. The
water was renewed once each day and the crabs were
fed with commercial pellets (Huaxu®, Xinxiang,
Henan Province, China) of 10% body weight. The
ingredients of the pellets include crud protein (≥30%),
crud fat (≥3%), crude ash (≥18%), total phosphorus
(≥1.0%), Ca (≥0.6%), lysine (≥1.3%) and water
(≤12.0%). After acclimated to the laboratory
conditions for 1 week, animals’ hepatopancreas,
eyestalk, gill, stomach, intestines, Y organ, heart and
claw muscle tissues were collected and immediately
stored at -80
0
C for cDNA cloning and expression
analysis. Moreover, the individuals in inter-molt stage,
identified according to the criterion we previously
reported (Tian et al. 2012) for starvation experiment.
Animals were reserved individually in different tanks
in order to avoid cannibalism. Hepatopancreas and
claw muscles were collected at the 0, 7 and 14 days
and stored at -80
0
C for RNA isolation and qRT-PCR.
2.2 Cloning of cDNA Encoding mLST8
in E. sinensis
Total RNA was extracted from hepatopancreas of E.
sinensis with TRIzol. RNA concentration and purity
were measured with ultraviolet spectrophotometer.
The cDNA was synthesized with Revert Aid kit
(Fermentas, USA) and used as PCR template later.
The PCR primers (Table 1) were designed based on
the TSA (transcriptome shotgun assembly) sequence
(GenBank accession no. GBZW01005746.1) in
NCBI database.
Table 1: The primers for mLST8 amplification in Eriocheir
sinensis.
Primer Sequence (5’−3’) Application
EsmLST8-F1 ATTACCTGTCT
TACCTGCC
RT-PCR
EsmLST8-R1 GTACCATCAC
CTCCTGTG
RT-PCR
EsmLST8-F2 CGCAGACTCC
CAACACATTA
qRT-PCR
EsmLST8-R2 GCTGTACTCG
CTCTTGATCTC
qRT-PCR
27S -F GGTCGATGAC
AATGGCAAGA
qRT-PCR
27S -R CCACAGTACT
GGCGGTCAAA
qRT-PCR
The amplification of EsmLST8 cDNA with PCR
in following system: 5.0μl cDNA template, 1.5μl
upstream and downstream primers (10 uM), 1.0μl
dNTP Mix (10 mM), 1.0μl Ex taq (Takara, Japan),
25.0μl 2×Ex taq Buffer (Takara, Japan), add sterile
deionized water to a total volume of 50 μl. The
reaction conditions were as follows: 94°C pre
denaturation for 2 min; 94°C denaturation for 30 sec,
55°C annealing for 30 sec, and 72°C extension for 1
min 40 sec, run 35 cycles; 72°C extension for 10 min.
The PCR products were detected by 1.2% agarose gel
electrophoresis, recovered by gel cutting, ligated into
the pUCm-T vector (Sangon, Shanghai), transformed
into E. coli and cultured in LB-Amp plates with X-gal
and IPTG. Positive colonies (white colonies) were
selected and sequenced with a 3730xl DNA Analyzer
(ABI) by Sangon Biological Co., Ltd. (Shanghai).
2.3 Bioinformatics Analysis of
EsmLST8
The ORF finder of the NCBI website and the translate
tool of Expasy were used to identify the reading frame
and translate it into amino acid sequences. Sequence
similarity analysis and multiple alignment were
performed with BLAST and COBALT (constraint-
based alignment tool for multiple protein sequences)
from NCBI as well as Clustal from Bioedit. The
editing of the cDNA and protein sequences was
carried out with Bioedit. CD-search from NCBI was
used for conserved functional domain prediction for
the protein (Marchler-Bauer et al., 2017), SWISS-
MODEL, Phyre2, PONDR and pyMOL software
(Mura et al., 2010) were used to predict and analyze
the structure and natural disorder regions of the
proteins. The CLUSTAL and MUSCLE multiple
alignment tools and the NJ (Neighbor-Joining) or
UPGMA tree building tool of MEGA-X were used to
build the cladogram of proteins (Kumar et al., 2018).
ABS 2022 - The International Conference on Agricultural and Biological Sciences
12
2.4 The Expression of EsmLST8 in
Different Tissues
The expression of EsmLST8 was evaluated by the 2-
ΔΔCt method with real-time PCR instrument
(Applied Biosystems® QuantStudio® 3). The cDNA
from different tissues was used as templates for qRT-
PCR with the primers designed based on the cDNA
sequence of ESmLST8 (Table 1). The 27s RNA was
used as an internal reference as it is the most stable
and suitable for E. sinensis (Huang et al., 2017). The
reaction was as follows: 95°C for 3 min; 40 cycles of
95°C for 5 sec, 60°C for 15 sec, and 95°C for 15 sec.
The melting curve generation from 65°C to 99°C in
steps of 0.5°C/s. Three technical replicates were
performed for qRT-PCR.
2.5 Statistical Analysis
Statistical analysis was performed using one-way
ANOVA with SPSS 18.0 software. Post hoc
comparisons of the means were performed using
Duncan’s least significant difference test with a
significance level of P ≤ 0.05. The values are
presented as the mean ± standard error (SE).
3 RESULTS
3.1 Sequence Analysis on cDNA and
the Deduced Protein of EsmLST8
The cDNA of 1001 nucleotides (nt) encoding a
putative EsmLST8 was cloned and sequenced, which
contains partial sequences of 5’- and 3’- untranslated
regions and the complete coding sequence (CDS) of
954 nt (Figure 1). The deduced 317 amino acids (aa)
sequence from the CDS was searched as query by
blast in non-redundant GenBank proteins database
(organisms were limited in crustaceans), the aligned
sequence with the highest score was target of
rapamycin complex subunit lst8-like in Penaeus
vannamei (GenBank accession no. XP027234568.1,
with the total score of 577 and the E-value of 0.0). We
designated the proteins as EsmLST8 (MTOR
associated protein, LST8 homolog in E. sinensis) and
performed further analysis.
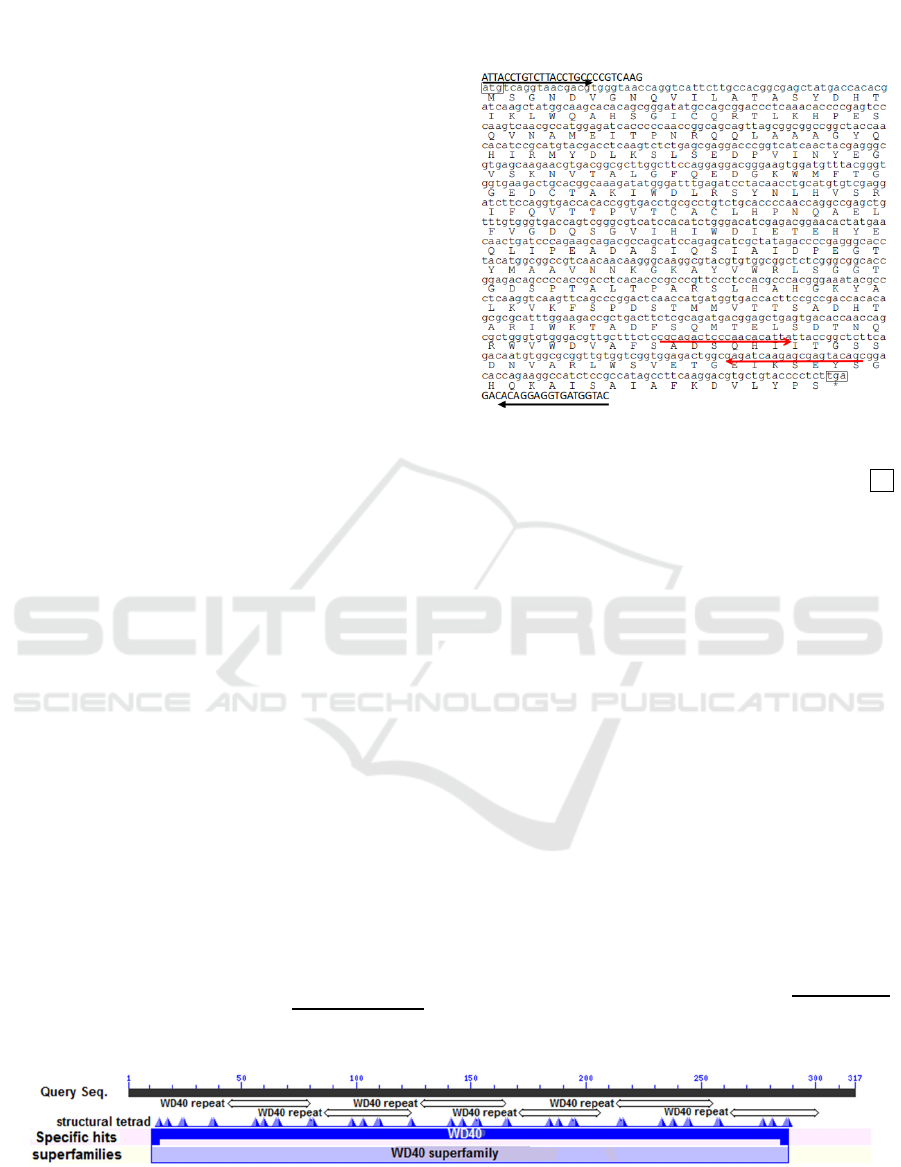
Figure 1: Nucleotide and deduced amino acid sequences of
the EsmLST8, including a partial 5’ and 3’ untranslated
region (UTR). The initiation codon is indicated with atg.
The stop codons are boxed and indicated with an asterisks.
The sequences corresponding primers for RT-PCR and
qRT-PCR are underlined with black and red arrows
respectively.
Predicated by the CD search tool, the EsmLST8
contains WD40 repeats structural motif, structural
tetrad on conserved domain WD40, covering 11-288
aa (Figure 2). A three-dimensional structure of
EsmLST8, which covered 10-312 aa, was predicted
using SWISS-MODEL workspace based on the
template (SMTL ID 5wbu.1.B) from human Crystal
structure of mLST8-PRAS40 complex (Yang et al.,
2017). The sequences of EsmLST8 and the template
human mLST8 (H-mLST8) used for modeling
showed 64.03% identity. The polypeptide backbones
of the EsmLST8 (depicted in red) and H-mLST8
(green) are almost perfectly overlapped, except that
Asp257-Trp262 of EsmLST8 form a shorter loop than
the counterpart region in H-mLST8 (Figure 3). The
EsmLST8 cDNA sequences have been deposited in
GenBank with the accession number of MN244302.
Figure 2: The conserved domains (WD40 domain) and function sites of the EsmLST8 covering 11-288 aa predicted by CD-
search.
Identification of MLST8, a Component of MTOR Complex 1 in Eriocheir Sinensis: CDNA Cloning and Its Expression Respond to
Starvation
13
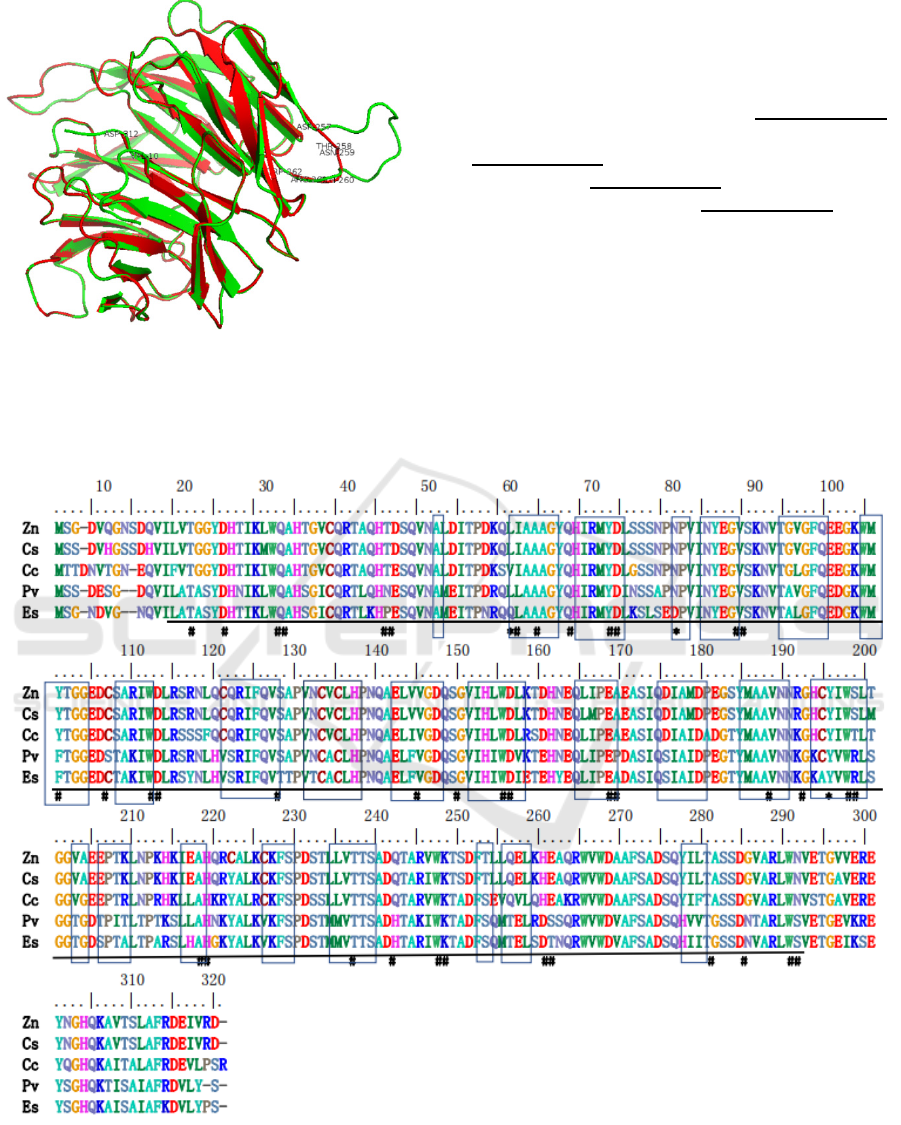
Figure 3: Predicted three-dimensional structure of the
EsmLST8 (red) covered 10-312 aa and its template human
mLST8(green). Val10, Asp312, Asp257-Trp262 were
labelled.
3.2 Multiple Sequence Alignment and
Phylogenetic Analysis on EsmLST8
EsmLST8 was aligned with homologs from P.
vannamei (GenBank accession no. XP027234568.1),
Zootermopsis nevadensis (GenBank accession no.
XP021941884.1), Cryptotermes secundus (GenBank
accession no. XP023707874.1) and Cephus cinctus
(GenBank accession no. XP015604398.1) using
COBALT on the NCBI web site. The sequence
identity of EsmLST8 compared with above 4
sequences are 0.854, 0.670, 0.664 and 0.668
respectively. The WD40 domain, WD40 repeat motif
and structural tetrad sites were showed in the
alignment. The function sites and structural motifs are
more conserved between two decapods or among
three insects respectively; While there are several
sites in EsmLST8 that are different with that in P.
vannamei as well as other 3 insects (Figure 4, marked
with the star symbol).
Figure 4: Multiple alignment of EsmLST8. Zn, Zootermopsis nevadensis; Cs, Cryptotermes secundus; Cc, Cephus cinctus;
Pv, Penaeus vannamei; Es, Eriocheir sinensis. Hashtags (#) indicate structural tetrad; Frames indicate W40 repeat motif
(structural motif); Underlines indicate WD40 domain; Stars (*) indicate different sites in Eriocheir sinensis compared with
Penaeus vannamei and other 3 insects.
ABS 2022 - The International Conference on Agricultural and Biological Sciences
14
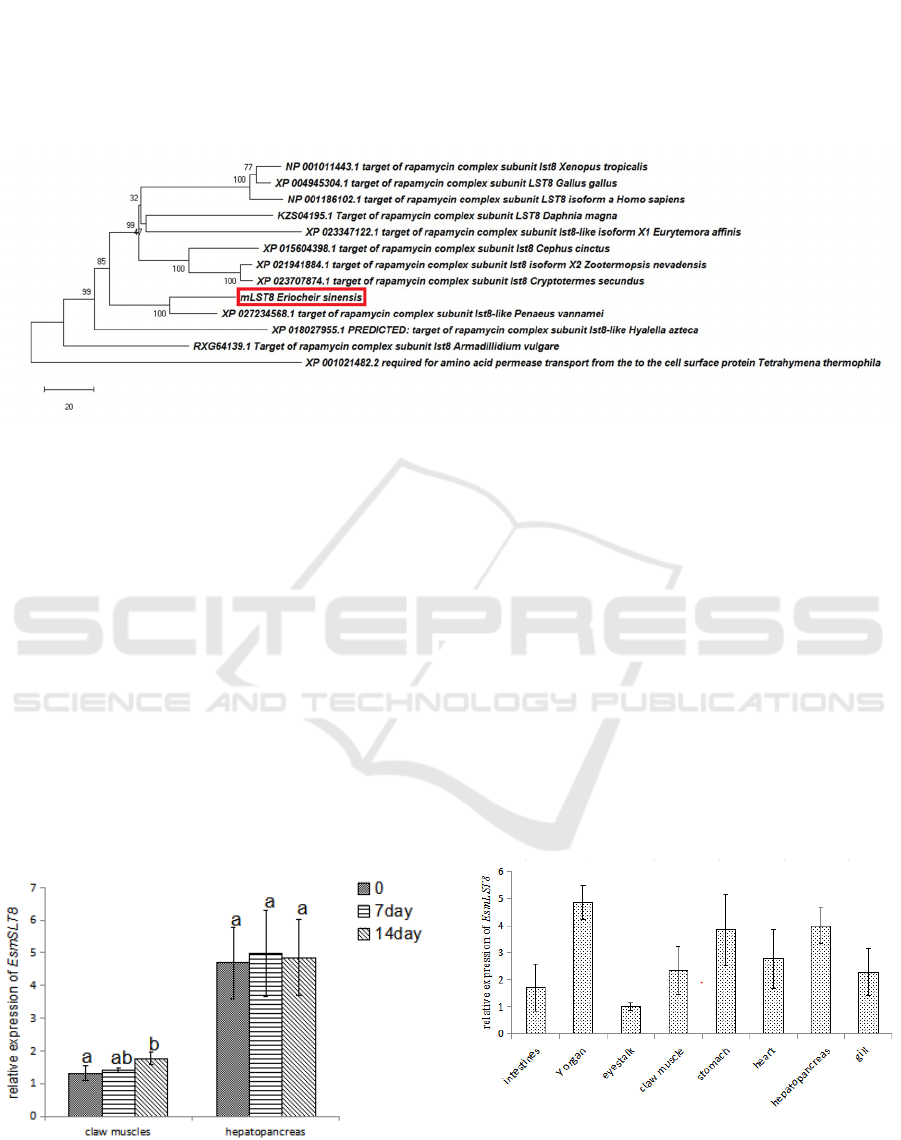
Phylogeny analysis together with homologs from
arthropods, vertebrates and a single cell species
(Tetrahymena thermophila) showed that EsmLST8
and mLST8 of P. vannamei clustered together, while
did not clustered with that from other crustaceans, i.e.
Daphnia magna, Eurytemora affinis, Hyalella azteca
and Armadillidium vulgare; compared with the 2
decapods, mLST8 in D. magna, E. affinis, Hyalella
Azteca and 3 insects (Cephus cinctus, Zootermopsis
nevadensis and Cryptotermes secundus) are more
close with that in 3 veterbrates (Homo sapiens, Gallus
gallus, and Xenopus tropicalis) (Figure 4).
Figure 5: Phylogenetic tree derived from multiple alignments of mLST8 (A) from different organisms. The evolutionary
history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa
clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with
branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary
distances were computed using the number of differences method and are in the units of the number of amino acid differences
per sequence. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.5). This analysis
involved 13 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
There was a total of 426 positions in the final dataset.
3.3 Tissue Distribution of mLST8 in
Juvenile E. sinensis
The expression pattern of EsmLST8 was examined
among the examined eight tissues (stomach,
hepatopancreas, intestines, eyestalk, Y organ, gill,
heart and claw muscles) from juvenile E. sinensis. It
was lowest expressed in eyestalk. The top three
tissues with high mRNA levels for the two genes are
Y organ, stomach and hepatopancreas (Figure 6).
Figure 6: The relative expression of mLST8 in different
tissues of juvenile E. sinensis, N=3.
3.4 The Expression of mLST8
Responded to Starvation in
Juvenile E. sinensis
Two weeks’ starvation had no significant effect on the
expression of EsmLST8 in hepatopancreas. While the
expression of EsmLST8 was increased significantly
at 14d in claw muscles after food deprivation (Figure
7).
Figure 7: The relative expression of mLST8 in different
tissues of juvenile E. sinensis, N=3. Different letters above
the errorbars indicate significant differences (P<0.05).
Identification of MLST8, a Component of MTOR Complex 1 in Eriocheir Sinensis: CDNA Cloning and Its Expression Respond to
Starvation
15

4 DISCUSSION
The mTOR signal pathway is highly conserved
among broad range organisms from single-celled
yeast to multiple-celled animals and plants. In the
yeast Saccharomyces cerevisiae, mLST8 homologue
negatively regulates amino acid biosynthesis as a
component of the mTOR pathway (Chen and Kaiser,
2003). In the plant Arabidopsis, mutations in the
homolog of mLST8 impaired plant growth, flowering,
and metabolic adaptation to long days (Moreau et al.,
2012). It is reported that mLST8 was upregulated in
human colon and prostate cancer cell lines and tissues,
and knocking down of its expression suppressed
formation of mTORC1/C2 complex, at the same time
inhibit tumor growth and invasiveness in different
human cancer cells (Kakumoto et al., 2015). Over
expression of mLST8 promoted normal epithelial
cells growth, while knockdown had no effect on their
growth (Kakumoto et al., 2015). We hypothesize that
in crustaceans the mLST8 is also important
components of the pathway and have important
functions involving in metabolic and growth
controlling. So, we cloned and sequenced the cDNA
encoding the proteins in E. sinensis and its expression
of transcription was investigated in this study.
The EsmLST8 contains the WD40 superfamily
domain including 7 WD40 repeats. The WD40 repeat
is a short structural motif of around 40 amino acids,
often terminating in a tryptophan-aspartic acid (W-D)
dipeptide (Neer et al., 1994). WD40 domain-
containing proteins have 4 to 16 repeating units, all of
which form a circularised beta-propeller structure
(Villanueva et al., 2016). The predicted structure of
EsmLST8 is consistent with this knowledge, and it is
highly conserved with and almost perfectly
superimposed to the human mLST8. The exception to
this is the region Asp257-Trp262 of EsmLST8, which
did not form the longer loop in the corresponding
region as human mLST8. What’s more, there are 3
sites with unique amino acids in regions of WD
repeats of EsmLST8 compared with mLST8 from P.
vannamei, C. cinctus, C. secundus, and Z. nevadensis.
The meaning of these differences on its function are
need to be investigated further in the future research.
The cDNA we cloned encoding an EsmLST8 was also
supported by the phylogenic analysis with mLST8
homologues from other organisms.
The EsmLST8 was expressed in all the examined
tissues. For crustaceans, the biosynthesis of
ecdysteroids in Y organ depends on mTOR pathway
(Abuhagr et al., 2016; Shyamal et al., 2018). However,
it was inhibited by MIH during the inter-molt stage of
the molting cycle. Accordingly, the transcripts
abundance of EsmLST8 was highest in Y organ and
lowest in eyestalk. Moreover, some much higher
expressions of EsmLST8 was also identified in
digestive organs, such as stomach and
hepatopancreas, indicating a strong digestion in
juvenile crabs contribute to their rapid growth is
connected with the mTOR signaling pathway.
Crustaceans often encounter starvation for a
short or long time in their livelihood due to different
reasons: molting, environmental changes, or
pollution (Hu et al., 2012). Hepatopancreas is a vital
organ for animal in growth and metabolism and store
much of lipids and glycogen in crustaceans (Tian et
al., 2012). During the inter-molt stage of decapods,
food deprivation causes the energy resources to be re-
allocated for tissue maintenance and survival
(Morales et al., 2012). Some decapods adopt an
adaptive strategy to avoid the usage of high costly
macromolecules instead preferentially utilize energy
from glycogen or lipid catabolism during food
deprivation period (Sacristan et al., 2017 ). This
contributes to maintain the stability of the proteins
and the intracellular pool of amino acids, which can
account for the stability of mTORC1 signaling, and
the transcriptions of EsmLST8 was kept around the
same level during the 14 days’ period of food
deprivation in hepatopancreas identified in these
studies.
However, the expression of EsmLST8 increased
in claw muscles under the conditions of 14 days’ food
deprivation in juvenile E. sinensi. This result is
consistent with the research report in species of
juvenile pacu, Piaractus mesopotamicus, in which a
short period of starvation induced the expression
increasing of anabolic genes, such as PI3K, mTOR,
mLST8 and RAPTOR in skeletal muscle (Paula et al.,
2017). In mouse and human cells, the activation of
mTORC1 was observed to promote an increase of
protein degradation as well as protein synthesis
(Zhang et al., 2014). Our results indicate that maybe
as well as in mammals, amino acids could be derived
from protein degradation in crustacean skeletal
muscles, which was triggered by food deprivation,
and maintained the intracellular pool of amino acids
for essential proteins synthesis via increasing the
expression of mTORC1 pathway components.
5 CONCLUSION
We cloned and sequenced the cDNA sequences
encoding an important mTOR pathway component
EsmLST8 in E. sinensis. EsmLST8 contains WD40
superfamily region comprising of WD40 repeats
ABS 2022 - The International Conference on Agricultural and Biological Sciences
16

domains, which make a circularized beta-propeller
structure. Y organ, stomach and hepatopancreas were
the top three tissues with high levels of EsmLST8
transcripts while the lowest expression of the gene is
in eyestalk. The transcriptional expression of
EsmLST8 was increased significantly at 14d in claw
muscles after two weeks’ food deprivation. The
results of this study provide basic data for studying
the roles of mTOR signaling pathway in the
regulation of crustacean growth and nutritional
metabolism.
REFERENCES
Abuhagr, A., Maclea, K., Chang, E., and Mykles, D. (2014).
Mechanistic target of rapamycin (mTOR) signaling
genes in decapod crustaceans: cloning and tissue
expression of mTOR, Akt, Rheb, and p70 S6 kinase in
the green crab Carcinus maenas, and blackback land
crab Gecarcinus lateralis. Comparative Biochemistry &
Physiology Part A Molecular & Integrative Physiology.
168: 25-39.
Abuhagr, A., MacLea, K., Mudron, M.., Chang, S., Chang,
E.,, and Mykles, D. (2016). Roles of mechanistic target
of rapamycin and transforming growth factor-beta
signaling in the molting gland (Y-organ) of the
blackback land crab, Gecarcinus lateralis. Comparative
biochemistry and physiology, Part A. Molecular and
integrative physiology 198A: 15-21.
Chen, E., and Kaiser, C. (2003). LST8 negatively regulates
amino acid biosynthesis as a component of the TOR
pathway. The Journal of Biophysical and Biochemical
Cytologyl. 161(2): 333-347.
Chen, J., and Long, F. (2018). mTOR signaling in skeletal
development and disease. Bone Res, 6: 1.
Das, S., Pitts, N.., Mudron, M.., Durica, D., and Mykles, D.
(2016). Transcriptome analysis of the molting gland (Y-
organ) from the blackback land crab, Gecarcinus
lateralis. Comparative Biochemistry & Physiology Part
D Genomics & Proteomics, 17: 26-40.
Das, S., Vraspir, L., Zhou, W., Durica, D., and Mykles, D.
(2018). Transcriptomic analysis of differentially
expressed genes in the molting gland (Y-organ) of the
blackback land crab, Gecarcinus lateralis, during molt-
cycle stage transitions. Comparative Biochemistry &
Physiology Part D Genomics & Proteomics, 28:37-53.
Hu, M., Tsang, S., Wang, Y., and Cheung, S.(2012). Effect
of prolonged starvation on body weight and blood-
chemistry in two horseshoe crab species:Tachypleus
tridentatus and Carcinoscorpius rotundicauda
(Chelicerata: Xiphosura). Journal of Experimental
Marine Biology & Ecology, 395(1-2): 112-119.
Huang, S., Chen, X., Wang, J., Chen, J., Yue, W., Lu, W.,
Lu, G., and Wang, C. (2017). Selection of appropriate
reference genes for qPCR in the Chinese mitten crab,
Eriocheir sinensis (Decapoda, Varunidae). Crustaceana,
90: 275-296.
Kakumoto, K., Ikeda, J., Okada, M., Morii, E., and
Oneyama, C. (2015). mLST8 Promotes mTOR-
Mediated Tumor Progression. PLoS One 10: e0119015.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K.
(2018). MEGA X: Molecular evolutionary genetics
analysis across computing platforms. Molecular
Biology Evolution, 35: 1547-1549.
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C.J.,
Lu, S., Chitsaz, F., Derbyshire, M.K., Geer, R.C.,
Gonzales, N.R., Gwadz, M., Hurwitz, D.I., Lu, F.,
Marchler, G.H., Song, J.S., Thanki, N., Wang, Z.,
Yamashita, R.A., Zhang, D., Zheng, C., Geer, L.Y., and
Bryant, S.H.(2017). CDD/SPARCLE: functional
classification of proteins via subfamily domain
architectures. Nucleic Acids Research, 45(D1): D200-
D203.
Morales, A., PeÂrez-JimeÂnez, A., FurneÂ, M., and
Guderley, H. (2012). Starvation, energetics, and
antioxidant defenses, in: D. Abele, J.P. VaÂzquez-
Medina, T. Zenteno-SavõÂn (Eds.), Oxidative stress in
aquatic ecosystems. Blackwell Publising, Oxford, 281-
294.
Moreau, M., Azzopardi, M., Clement, G., Dobrenel, T.,
Marchive, C., Renne, C., Martin-Magniette, M.L.,
Taconnat, L., Renou, J.P., Robaglia, C., and Meyer, C.
(2012). Mutations in the Arabidopsis homolog of
LST8/GbetaL, a partner of the target of Rapamycin
kinase, impair plant growth, flowering, and metabolic
adaptation to long days. Plant Cell, 24: 463-481.
Mura, C., McCrimmon, C.M., Vertrees, J., and Sawaya,
M.R. (2010). An introduction to biomolecular graphics.
PLOS Computational Biology, 6(8):e1000918.
Mykles, D.(2011). Ecdysteroid metabolism in crustaceans.
The Journal of Steroid Biochemistry and Molecular
Biology, 127: 196-203.
Neer, E., Schmidt, C., Nambudripad, R., and Smith, T.
(1994). The ancient regulatory-protein family of WD-
repeat proteins. Nature, 371: 297-300.
Paula, T., Zanella, B., Fantinatti, B.., Moraes, L., Duran, B.,
Oliveira, C., Salomao, R.., Silva, R.., Padovani, C.,
Santos, V.., Mareco, E., Carvalho, R., and Dal-Pai-Silva,
M. (2017). Food restriction increase the expression of
mTORC1 complex genes in the skeletal muscle of
juvenile pacu (Piaractus mesopotamicus). PLoS One,
12(5): e0177679.
Sacristan, H., Rodriguez, Y., De Los Angeles Pereira, N.,
Lopez Greco, L.., Lovrich, G., and Fernandez Gimenez,
A. (2017). Energy reserves mobilization: Strategies of
three decapod species. PLoS One, 12(9): e0184060.
Saxton, R., and Sabatini, D. (2017). mTOR Signaling in
growth, metabolism, and disease. Cell, 169(2): 361-371.
Shyamal, S., Das, S., Guruacharya, A., Mykles, D.L., and
Durica, D.S. (2018). Transcriptomic analysis of
crustacean molting gland (Y-organ) regulation via the
mTOR signaling pathway. Scientific Reports, 8(1):
7307.
Tian, Z., Kang, X., and Mu, S. (2012). The molt stages and
the hepatopancreas contents of lipids, glycogen and
selected inorganic elements during the molt cycle of the
Identification of MLST8, a Component of MTOR Complex 1 in Eriocheir Sinensis: CDNA Cloning and Its Expression Respond to
Starvation
17

Chinese mitten crab Eriocheir sinensis. Fisheries.
Science,78(1): 67-74.
Villanueva, M.., Islas-Flores, T., Ullah, H., and Gilroy, S.
(2016). Editorial: Signaling through WD-Repeat
Proteins in Plants. Frontiers in Plant Science,, 7:1157.
Webster, S.., Keller, R., and Dircksen, H. (2012). The CHH-
superfamily of multifunctional peptide hormones
controlling crustacean metabolism, osmoregulation,
moulting, and reproduction. General & Comparative
Endocrinology,175(2): 217-233.
Yang, H., Jiang, X., Li, B., Yang, H.J., Miller, M., Yang, A.,
Dhar, A., and Pavletich, N.P. (2017). Mechanisms of
mTORC1 activation by RHEB and inhibition by
PRAS40. Nature, 552: 368-373.
Yang, H., Rudge, D., Koos, J., Vaidialingam, B., Yang, H.,
and Pavletich, N.(2013). mTOR kinase structure,
mechanism and regulation. Nature, 497: 217-223.
Yonezawa, K., Tokunaga, C., Oshiro, N., and Yoshino, K.
(2004). Raptor, a binding partner of target of rapamycin.
Biochemical and Biophysical Research
Communications, 313: 437-441.
Zhang, Y., Nicholatos, J., Dreier, J.R., Ricoult, S.J.,
Widenmaier, S.B., Hotamisligil, G.S., Kwiatkowski,
D.J., and Manning, B.D.(2014). Coordinated regulation
of protein synthesis and degradation by mTORC1.
Nature, 513:440-443.
ABS 2022 - The International Conference on Agricultural and Biological Sciences
18
