
The Secondary Structure Analysis and Protein Identification of
Esterase Were Performed by Circular Dichroism
Xinzhi Cao
1*
, Liming Zhong
1
, Shuyuan Li
2
, Anqi Zhou
1
, Kaizheng Zhang
1
and Zeli Song
1
1
Department of Bioengineering, Sichuan University of Science and Engineering, Zigong, Sichuan 643000, China
2
Wuliangye Group Co. Ltd., Yibin 644007, China
Keywords:
Esterifying Enzyme, Protein Structure, Round Two Chromatographic.
Abstract:
In order to improve the yield of esterase and achieve the purpose of improving liquor quality rate and
aroma, the structure of esterase was studied by circular dichroism method and the protein was identified by
liquid mass spectrometry (LC-MS/MS). The results showed that the content of esterification enzyme Helix
(Helix) was 17.10%, anti-parallel β -folding structure was 29.70%, parallel β -folding structure was 5.40%,
β -rotation structure was 18.60%, irregular coil structure was 37.80%. This indicates that esterase is a kind
of protein with irregular curl. The β -folding structure of esterase accounted for 35.10%, indicating that the
secondary structure of esterase had a certain rigidity, but the β -rotation and random curl structure accounted
for 56.40%, indicating that the various residues in esterase peptide segment had a large degree of freedom,
good flexibility, but poor stability. Protein identification results showed that RHICH Lipase peptide had the
highest content, 906, coverage rate of 76.35%, total theoretical amino acids of 389 protein. The number of
PSEFL DNA-Binding Response regulator peptide was 14, the coverage rate was 32.93%, and the total
number of theoretical amino acids was 246. Then, the number of BURPL 50S ribosomal protein L5 peptide
was 10, the coverage was 18.99%, and the total number of theoretical protein amino acids was 179; Then
there was 9GAMM 30S ribosomal protein S19 peptide, the number was 7, the peptide coverage was 15.22%
and the total number of theoretical amino acids was 92; Finally, the peptide of 9GAMM Succinate
dehydrogenase iron-sulfur subunit was 7, the coverage rate was 16.03, and the total number of protein
theoretical amino acids was 237.
1 INTRODUCTION
Esterase (esterase.C.3.1.1.1), also known as
carboxyl esterase, is an enzyme that can hydrolyze
carboxyl ester bonds and catalyze the synthesis of
low grade fatty acid esters (Chen, 2017), which has
the ability to catalyze ester synthesis and
decomposition. Therefore, liquor industry is used to
call it esterase or ester decomposition enzyme.
Protein refers to the polymer formed by the
connection of 20 different amino acids (Yao, 2006;
Xu, 2011; S Sirén, 2020) The structure of protein
includes the chemical structure and spatial structure
of protein (Xu, 2020; Gao, 2010). The methods
(Zhou, 2021; Wei, 2021) to study the secondary
structure and advanced structure of protein include
X-ray crystal diffraction technology, nuclear
magnetic resonance technology and circular
dichroism technology (Xiao, 2020). But the first two
methods are limited by many factors, it is difficult to
analyze. The analytical scanning of circular
dichroism spectrometer plays an important role in
studying the secondary structure and advanced
structure of proteins, which is a special absorption
spectrum (Huang, 2019). The circular dichroism
spectrum of biological macromolecules such as
proteins detected by circular dichroism
chromatography is used to obtain the secondary
structure of biological macromolecules (Tong,
2018). Therefore, CIRCULAR dichroism is widely
used in protein folding and conformation research
(Liu, 2012).
This paper uses circular dichroism to analyze the
secondary structure of esterase and identify the
esterase protein to provide a theoretical basis for the
application of esterase in wine industry.
76
Cao, X., Zhong, L., Li, S., Zhou, A., Zhang, K. and Song, Z.
The Secondary Structure Analysis and Protein Identification of Esterase Were Performed by Circular Dichroism.
DOI: 10.5220/0012002200003625
In Proceedings of the 1st International Conference on Food Science and Biotechnology (FSB 2022), pages 76-79
ISBN: 978-989-758-638-5
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
2 MATERIALS AND METHODS
2.1
Strains and Chemical Reagents
Staphylococcus aureus: staphylococcus aureus was
isolated and screened from Wuliangye Daqu, and the
strain producing esterification enzyme was
identified as STaphylococcus aureus according to the
morphological characteristics of the fungus colony
and molecular biology.
Acetonitrile, formic acid and ammonium
bicarbonate were all ms grade. Dithiothreitol and
iodoacetamide were analytically pure. Trypsin
sequencing grade.
2.2
Circular Dichroism Detection
The sample was dissolved in water at a
concentration of 0.2 ug /uL. The initial wavelength
was set at 180 nm, the end wavelength was set at
260 nm, the step size was 1 nm, the collection time
was 1 s/ point, and the cuvette width was 0.1 cm.
The blank control solution was sampled with 300 uL
and deducted after measurement. Then test the
sample with 300 uL, and save the data after test.
2.3
Protein Identification
2.3.1 LC - MS/MS Detection
Packed with Acclaim PepMap RPLC C18, 5 μm,
100A; 150 μm I.D. × 150 mm, Packed with Acclaim
PepMap RPLC C18, 1.9 μm, 100A; Mobile phase A:
0.1% formic acid; Mobile phase B: 0.1% formic
acid, 80% ACN; Flow rate: 600 nL/min; Analysis
time of each component: 60 min.
2.3.2 Mass Spectrometry Conditions
Primary mass spectrometry parameters: Resolution:
70,000; AGCtarget: 3 e6; MaximumIT: 100 ms;
Scanrange: 300 to 1400 m/z.
Secondary mass spectrometry parameters:
Resolution: 17,500; AGCtarget: 1 e5; MaximumIT:
50 ms; TopN: 20; An NCE/steppedNCE: 28.
2.3.3 Database Search
The mass spectrometry raw file retrieves the target
protein database using Byonic.
3 RESULTS AND DISCUSSION
3.1
CD Spectrum of Esterase in Far
ULTRAVIOLET Region
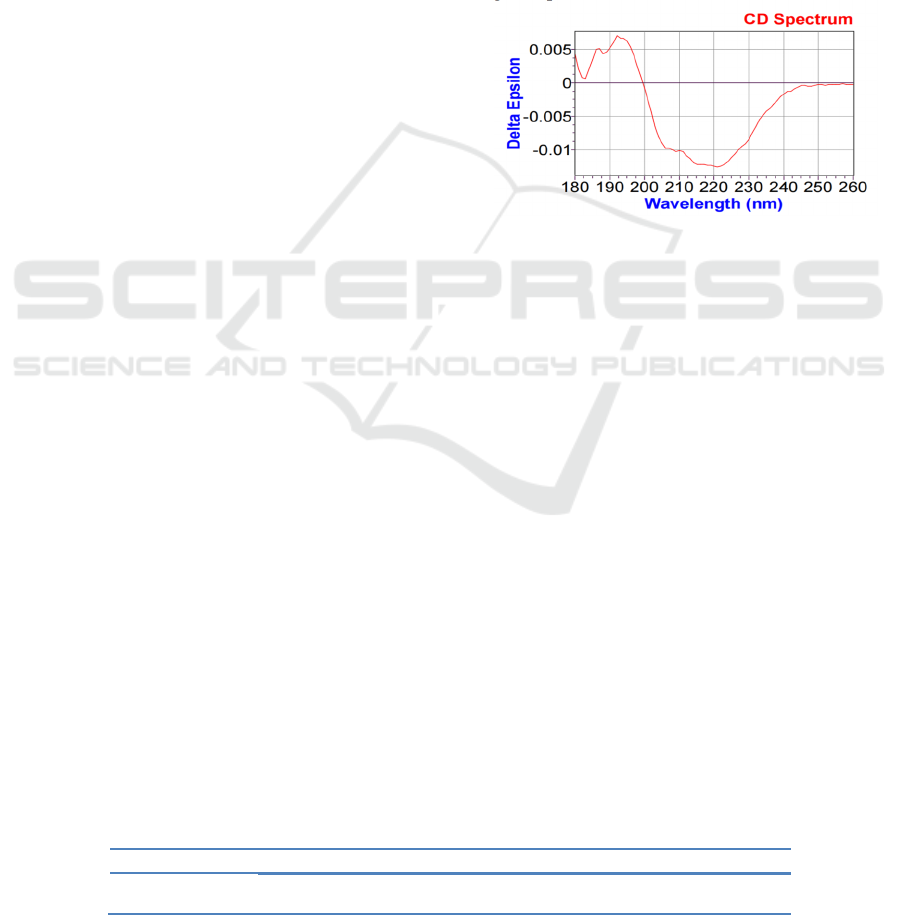
The UV CD spectrum of esterification enzyme was
shown in Fig.1. According to Fig.1, there was a
positive polarization peak at 195 nm and a small
shoulder at 187 nm, while the control group had a
wide negative shoulder at 210 nm and a wide
negative peak at 220 nm.
The abscissa represents the scanning wavelength and the
ordinate represents the ellipticity
Figure 1: Far-ultraviolet (180-260 nm) scanning of
esterification enzyme.
3.2
Secondary Structure of
Esterification Enzyme
The secondary structure of the sample was fitted
with CDNN software, including Helix, Antiparallel,
Parallel, beta-turn and RNDM.coil.
The analysis results were as follows:
esterification enzyme helix structure 17.10%,
antiparallel structure 29.70%, parallel β -folding
structure 5.40%, β -rotation structure 18.60%,
random coil structure 37.80%. This indicates that
esterase is a kind of protein with irregular curl.
3.3
Esterase Protein Identification
Raw files generated by esterase through LC-MS/MS
data collection were opened with Xcalibur, and total
ion flow chromatography fig. 2 could be seen as
follows:
Table 1: Calculation of secondary structure ratio in esterification enzyme.
The sam
p
le Helix Anti
p
arallel Parallel Beta-Turn Rndm. Coil
Esterifying enzyme 17.10% 29.70% 5.40% 18.60% 37.80%
The Secondary Structure Analysis and Protein Identification of Esterase Were Performed by Circular Dichroism
77
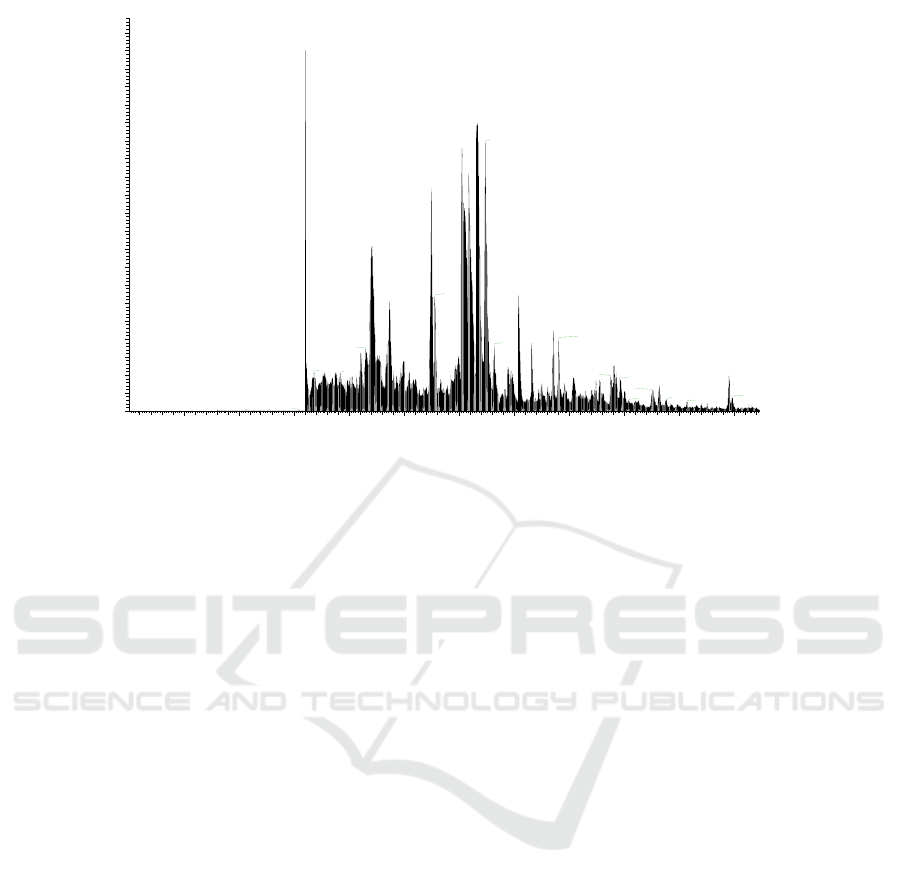
Figure 2: Total ion flow chromatogram of esterification enzyme.
The results of protein identification were
retrieved by software Byonic database as shown in
the attached page:
From the attached pages: The results showed that
the top ten proteins with high scores were RHICH
Lipase, PSEFL DNA-Binding Response regulator,
BURPL 50S ribosomal protein L5 and 9GAMM 30S
Ribosomal protein S19, 9GAMM Succinate
dehydrogenase iron-sulfur subunit, 9GAMM
Succinate--CoA ligase [ADP-forming] Subunit beta,
ARHIZD Lipase, BURPL 50S Ribosomal protein
L14, 9GAMM 2-methylisocitrate lyase, 9GAMM
Sulfate Adenylyl transferase subunit 2. The number
of RHICH Lipase peptide was 906, the coverage rate
was 76.35%, and the total number of theoretical
amino acids was 389. The number of peptide in
PSEFL DNA-Binding Response regulator was 14,
the coverage rate was 32.93%, and the total number
of theoretical amino acids was 246. Then the
number of peptides in the BURPL 50S Ribosomal
protein L5 was 10, the coverage was 18.99%, and
the total number of theoretical amino acids in the
protein was 179; Then 9GAMM 30S ribosomal
protein S19 had a peptide number of 7 and a peptide
coverage of 15.22% with a theoretical total of 92
amino acids; Fifth, the number of peptide of
9GAMM Succinate dehydrogenase iron-sulfur
subunit was 7, the coverage rate was 16.03, and the
total number of protein theoretical amino acids was
237; The sixth score was 9GAMM Succinate-- the
maximum number of peptides in CoA ligase
[ADP-forming] subunit beta was 2, the coverage
rate was 7.22%, and the total number of theoretical
amino acids in protein was 388. In the seventh place,
ARHIZD Lipase had a maximum of 16 peptides, a
coverage rate of 9.77%, and a total of 389
theoretical amino acids; For the eighth BURPL 50S
ribosomal protein L14, the maximum number of
peptides was 9, the coverage rate was 22.95%, and
the total number of theoretical amino acids was 122;
The number of 9GAMM 2-methylisocitrate lyase is
3, the coverage rate is 5.19%, and the total number
of protein theoretical amino acids is 289; The
number of peptides in 9GAMM Sulfate adenylyl
transferase subunit 2 is 3 at most, the coverage rate
is 8.50%, and the total number of theoretical amino
acids in protein is 306.
4 CONCLUSIONS
Using circular dichroism to analyze esterase, the β
-fold structure of esterase accounted for 35.10%, β
-rotation and random curl structure accounted for
56.40%, indicating that esterase is a protein mainly
with random curl, and each residue in esterase
peptide segment has a large degree of freedom, good
flexibility, poor stability. Protein identification
results showed that RHICH Lipase had the highest
protein content, with 906 peptides, 76.35% coverage
rate and 389 theoretical amino acids. PSEFL
DNA-Binding Response Regulator had 14 peptides,
the coverage rate was 32.93%, and the total number
of theoretical amino acids was 246. Then, the
number of peptide fragments in the BURPL 50S
RT: 0.04 - 57.36
5 10 15 20 25 30 35 40 45 50 55
Time (min)
0
5
10
15
20
25
30
35
40
45
50
55
60
65
70
75
80
85
90
95
100
105
Relative Abundance
16.04
31.66
32.40
30.27
27.50
22.10
35.4327.84
23.70
38.60
39.08
33.18 36.62
21.54
24.98
44.12
16.84
19.29
54.56
43.81
40.46
44.65
48.21
47.56
54.79
48.85
45.43
50.74
56.64
8.06
15.52
12.8210.84
NL:
3.24E10
TIC MS
化
FSB 2022 - The International Conference on Food Science and Biotechnology
78

Ribosomal protein L5 was 10, the coverage was
18.99%, and the total number of theoretical amino
acids in the protein was 179. Then the number of
peptides in 9GAMM 30S ribosomal protein S19 was
7, the coverage rate was 15.22%, and the total
number of theoretical amino acids in protein was 92.
Finally, the peptide number of 9GAMM Succinate
dehydrogenase iron-sulfur subunit was 7, the
coverage rate was 16.03, and the total number of
protein theoretical amino acids was 237. Different
methods and molecular mechanisms are used to
change the properties of esterification enzyme.
Therefore, protein identification combined with
esterification enzyme can provide ideas for the
experimental design of improving the properties of
esterification enzyme, so as to promote the
high-quality development of wine industry.
AUTHOR INFORMATION
Xinzhi Cao (1965-), male, professor, ph. D., mainly
engaged in food biotechnology, E-mail addresses:
caoxinzhi@163.com
ACKNOWLEDGMENTS
Fund Project: Cooperation project of Wuliangye
Yibin Co., Ltd (CXY2019ZR012
REFERENCES
Chen Zhongwei, WANG Keke, LIU Yanxia, et al. Effects
of inorganic salts on the structure and activity of lipase
in Wheat germ [J]. Journal of Cereals and Oils of
China, 2017, 32(6):7.
Gao Ying, Ye Xiaoli, Li Xuegang, et al. Extraction of
Polygonum flavum polysaccharide and its inhibitory
effect on α -glucosidase [J]. Chinese Patent Medicine,
2010, 32(12):5. (in Chinese).
HUANG W Q. Study on the enzymatic properties and
structure and function relationship of Aspergillus
fumigatus lipase AFLB [D]. South China University
of Technology, 2019.
Liu Y. Screening and identification of two
lipase-producing strains and cloning and expression of
lipase gene [D]. Central China Normal University,
2012.
Tong C D. Study on structure and function of
R-2-halogenated acid dehdiv-R and lipase PaL [D].
Zhejiang University, 2018.
S Sirén, Dahlstrm K M, Puttreddy R, et al. Candida
antarctica Lipase A-Based Enantiorecognition of a
Highly Strained 4-Dibenzocyclooctynol (DIBO) Used
for PET Imaging[J]. Molecules, 2020, 25(4).
WEI Zixiang, ZHANG Liuqun, Lei Lei, et al. Rational
design of lipase of Thermomyces lanuginosus to
improve its activity and temperature stability [J].
Chinese Journal of Bioengineering, 2021, 41(2):7.
Xu Qiang, Yang Tiqiang, Wang Miao. Effect of vacuum
on secondary structure of superoxide dismutase by
circular dichroism spectroscopy [J]. Vacuum, 2011,
48(3):4.
Xu Haoxi, LIU Lei. Research Status and direction of
protein structure analysis [J]. Journal of Tongling
University, 2020, 19(5):4.
Xiao Zhigang, Yang Guoqiang, Yang Qingyu, Wang
Lishuang, Zhang Xueping, Guo Shilong, Li Zhe, Yang
Shu. Structure and antioxidant activity of phospholipid
of linolenic acid synthesized by enzymatic method [J].
Food Science, 2020, 41(22):7.
Yao Zhanquan, Aodun Gerele, Xu Qiang, et al. Effect of
electric field on secondary structure of α -amylase by
circular dichroism spectroscopy [J]. Chinese Journal
of Analytical Measurement, 2006, 25(4):4.
Zhou Yanjie, Geng Peng, Shi Yi, et al. Recombination
expression and enzymatic properties of thermophilic
lipase from Rhodococcus rosiophilus [J]. Food and
Fermentation Industries, 2021, 47(13):7.
The Secondary Structure Analysis and Protein Identification of Esterase Were Performed by Circular Dichroism
79
