
Methods for Clinical Disease Classification and Clustering: A Review
Yong Shuai
1,2,*,†
, Xiaoxia Dai
1,3,†
, Xiaodong Wang
1
, Zeshan Liang
1,3
, Gang Yan
1,3
and Ming Chen
1,3
1
hongqing CEPREI Industrial Technology Research Institute Co., Ltd, 401332 Chongqing, China
2
Chongqing Key Laboratory of Reliability Technologies for Smart Electronics, 401332 Chongqing, China
3
CEPREI Innovation (Chongqing) Technology Co., Ltd, 401332 Chongqing, China
Keywords: Clinical Classification, Clustering, Feature Selection, Review.
Abstract:
The appropriate use of accurate clinical classification is of great importance for appropriate clinical diagnosis,
treatment, and for meaningful clinical research. Current clinical classification methods lack unified norms and
methods in feature selection, number of classification, and classification methods. It has thus become
necessary to discuss and critique current methods of clinical classification, and to discuss and formulate
constructive opinions regarding clinical classification in the future. We conducted a literature review of open
source references containing predetermined terms published in Chinese and English from 2003 to 2021. Our
search retrieved 59 studies concerning classification methods among different diseases. General processes,
feature selection, and classification methods of clinical classification were then summarized and analyzed.
The existing problems of current literature in clinical classification data sources, feature selection, number of
classification, and methods were analyzed. We then propose targeted measures with respect to these problems,
to help researchers find a suitable method for classification. Through a literature review, we have discovered
the shortcomings of current clinical classification methods, and herein suggest corresponding
countermeasures. We hope to improve the scientific basis of future classification methods and the
interpretability of classification results by implementation of the countermeasures proposed in this review, so
that classification results can be more universally recognized, and used over a wider range.
1 INTRODUCTION
In the process of analyzing clinical disease data, as
the clinical manifestations of disease become
relatively atypical and lacking in specificity,
researchers usually classify patients based on one or
more clinical features and thus conduct research on
different categories of patients, in order to enable
clinicians to gain a deeper understanding of the
characteristics and pathological changes of diseases,
and to execute targeted diagnosis and treatment. This
process is called clinical classification. In the process
of China's response to the COVID-19 epidemic,
accurate classification has reduced the mortality rate
and severe disease rate, and improved the cure rate,
which showed that clinical classification was of great
significance. (CDC, 2020)
Science-based clinical classification not only
helps doctors focus on the specific disease-related
*
Corresponding author
†
Yong Shuai and Xiaoxia Dai contributed equally to this
work and should be considered co-first authors
changes of different types of patients and formulate
appropriate treatment plans, but also assists doctors in
accurately assessing the patient's disease evolution
and prognosis. Especially for medical staff in remote
areas and resource-limited countries, and in regions
with insufficient medical resources, scientific
classification results can effectively guide the
diagnosis and treatment of patients, so as to
accurately control the patient's condition, improve the
patient's prognosis, and reduce the rate of clinical
deterioration and mortality.
Taking current ideas and methods of clinical
classification into account, this paper summarizes the
general clinical classification process, feature
selection methods, and classification methods,
discusses the existing problems in current clinical
classification methods, and proposes
countermeasures to these problems. The purpose of
this review is to help improve the scientific basis of
116
Shuai, Y., Dai, X., Wang, X., Liang, Z., Yan, G. and Chen, M.
Methods for Clinical Disease Classification and Clustering: A Review.
DOI: 10.5220/0012014700003633
In Proceedings of the 4th International Conference on Biotechnology and Biomedicine (ICBB 2022), pages 116-124
ISBN: 978-989-758-637-8
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
clinical disease classification methods, to enhance the
interpretability of clinical classification results, and to
allow more researchers to find consensus with
classification results of different datasets.
2 METHOD
Literature data was obtained through the following
websites: https://pubmed.ncbi.nlm.nih.gov and
https://www.cnki.net. Search keywords included
'clinical', 'feature selection', 'classification', and
'cluster'. The search language for
https://pubmed.ncbi.nlm.nih.gov (Pubmed) was
English, and the search languages for
https://www.cnki.net/ (CNKI) were Chinese and
English.
In the literature review, we focused on the feature
selection methods and classification methods of each
literature item, and conducted a comparative analysis.
Since there are currently more than 300,000 Chinese
and English documents related to clinical
classification and clustering, after screening the
content of the title, abstract, and methods, based on
the authors' understanding of these papers, we
included 59 articles for our review published in
English and Chinese from July 2003 to September
2021.
3 RESULTS
Through literature review, we evaluated the general
process of clinical classification, and summarized
feature selection methods and clinical classification
methods. The relevant results were as follows.
3.1 Summary of General Clinical
Classification Process
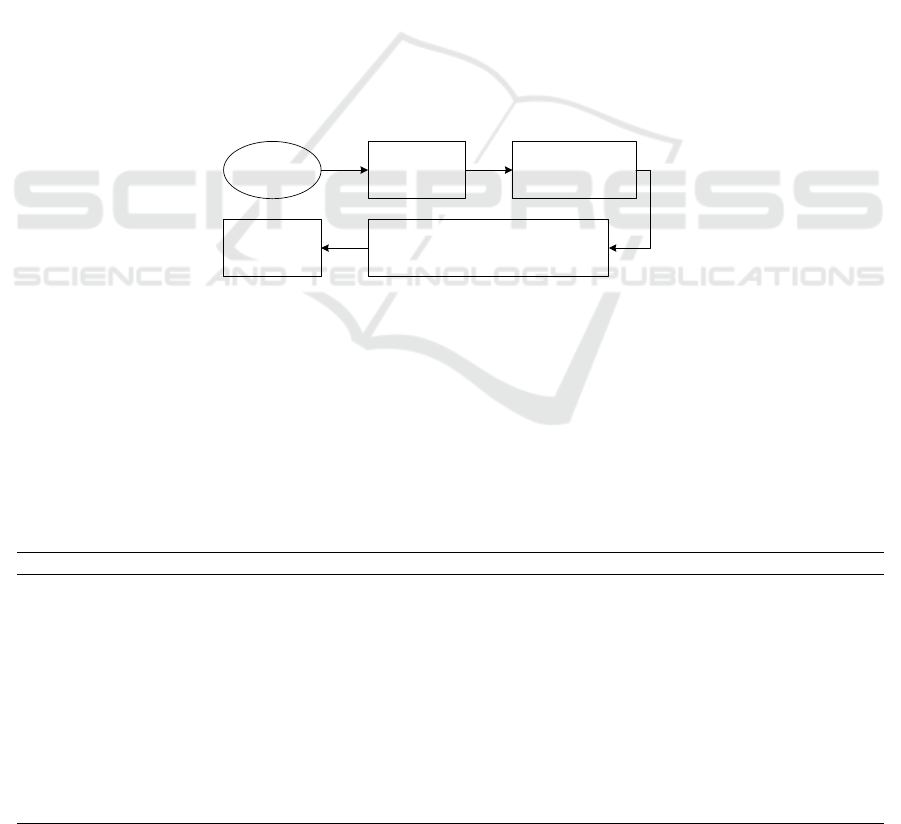
Based on the literature review, we found that clinical
classification can be understood to be a rigorous data
analysis process, and its general classification process
is shown in Figure 1. The modeling process of the
clinical classification process includes data source
preparation, feature selection, classification method
design, statistical analysis and interpretation of
classification results, and knowledge archiving.
Feature
selection
Data source
Classification or
clustering
Clinical analysis of
classification/clustering results
Knowledge
archives
Figure 1: General flow chart of clinical classification.
Generally, data preparation (Ahady, 2021; Shuai,
2018, Sun, 2020) included data collection, data
integration, data cleaning, data conversion, and data
specification, among which data cleaning, data
conversion, and data specification are collectively
referred to as data preprocessing. Since both data
preparation and statistical analysis have standard
research methods, we then reviewed the feature
selection and classification method design by the
general flow chart of clinical classification. The
methods for feature selection and classification
method extracted from the 59 studies are showed in
Table 1.
Table 1: Methods for feature selection and classification method extracted from 59 studies.
Total Numbe
r
Reference Numbe
r
Feature selection
Feature selection based on consensus or professional
experience
55 4-55, 61, 62, 63
Feature selection based on risk factor analysis 2 56,57
Feature selection based on the machine learning model 2 59-60
Clinical
classification
methods
Standard or consensus-based classification methods 7 4-7,21,23,61
Personal professional knowledge and clinical
experience-based classification methods
31
8-13, 18-20, 22, 24-
40, 56, 57, 62, 63
Supervised learning classification model-based
classification methods
6 14-16, 55, 59, 60
Unsupervised learning clustering model-based
classification methods
15 17, 41-54
Methods for Clinical Disease Classification and Clustering: A Review
117

3.2 Feature Selection
It is generally accepted that a patient generates a
substantial volume of disease-related data, including
demographic information, physical examination
information, auxiliary examination information,
clinical disease characteristics, pathological change
information, treatment information, prognostic
information, etc. These data contain hundreds of
individual features. Since the selection of different
features directly affects the classification results,
precisely which features are to be selected for clinical
classification is an important step in the clinical
classification of patients.
In current literature, feature selection methods for
clinical classification mainly include 3 types, i.e.,
feature selection based on consensus or professional
experience, feature selection based on risk factor
analysis, and feature selection based on a machine
learning model.
(1) Feature selection based on consensus or
professional experience
Most clinical classification literature (Chen, 2020;
Xiang, 2009; Shi, 2010) did not clearly indicate
specific feature selection methods. The features used
for clinical classification in these documents were
used directly, without specific reasons for their use
being given. We can think of the method used here to
select features as based on the author's personal
understanding of the disease or their clinical
experience.
(2) Feature selection based on risk factor analysis
Risk factor analysis included risk factors for
mortality (Yuan, 2020) and risk factors for prognosis
(Wang, 2020). The basic idea underlying this method
was to use single factor analysis and logistic
multivariate regression analysis to find features that
could be used to classify patients.
(3) Feature selection based on the machine
learning model
The feature selection algorithm is a machine
learning model that can reduce the complexity of a
problem and improve the accuracy, robustness, and
interpretability of the algorithm (Li, 2019). Feature
selection models used in clinical classification mainly
include Recency Frequency Engagements (RFE)
(Noor, 2015) and the SelectKBest method, based on
the Chi-Squared test (Li, 2020).
3.3 Clinical Classification Methods
After the features have been selected, the authors of
these articles then use these features to carry out
clinical classification. The main classification
methods can be regarded as standard or consensus-
based classification methods, personal professional
knowledge and clinical experience-based
classification methods, supervised learning
classification model-based classification methods,
and unsupervised learning clustering model-based
classification methods.
(1) Standard or consensus-based classification
methods
Standard or consensus-based classification
methods mainly use guidelines published by
professional institutions (CDC, 2020), diagnosis and
treatment standards (Thijs, 2010; Viprakasit, 2018;
Jessica, 2018), expert consensus statements (Chen,
2020), and traditional medicine diagnosis and
treatment rules (such as Tibetan medicine (Hua,
2020) and traditional Chinese medicine (Tian, 2021)
to carry out clinical classification.
Since guidelines published by professional
institutions, diagnosis and treatment standards, expert
consensus, and traditional medical diagnosis and
treatment rules are based on the sum of the
professional knowledge system of most authoritative
experts, this clinical classification method can be
regarded as a method based on expert knowledge.
(2) Personal professional knowledge and clinical
experience-based classification methods
The personal professional knowledge and clinical
experience-based classification methods rely on the
professional knowledge and understanding of the
disease by individual doctors and researchers. This
clinical classification method often uses statistical
analysis on one or more feature values to achieve
classification.
This method is more commonly used in current
literature articles. Since many diseases do not have a
standard classification method, many authors of
articles select some clinical or other feature to classify
patients based on personal professional knowledge,
experience, and their understanding of the disease.
Because this classification method involves high
subjectivity, different professionals may generate
different classification results.
Currently, personal professional knowledge and
clinical experience-based classification methods use
clinical manifestations or symptoms (including
clinical, pathophysiological mechanism, pathological
features, anatomy, pathology, and patient status)
(Monica, 2020), treatment programs (Sun, 2013),
genome sequencing results (Sandra, 2018), and non-
clinical medical conditions (such as medical
insurance (Xiang, 2009), and distinction between
inpatients and outpatients (Shi, 2010) to classify
patients.
ICBB 2022 - International Conference on Biotechnology and Biomedicine
118

(3) Supervised learning classification model-
based classification methods
The supervised learning classification algorithm
is an algorithm that establishes independent reference
standards in the labeled training data, built
classification models (such as support vector machine
(SVM), logistic regression (LR), etc.), and classifies
new data on this basis.
Current clinical classification methods based on
supervised learning models include SVM (Jonathan,
2016), neural networks with principal component
analysis (Ahmad, 2020), decision tree analysis
(Ahmad, 2020), elastic net (Ahmad, 2020) random
forest (Varol, 2020), multilayer perceptron (Varol,
2020), and extreme gradient boosting (Ma, 2020).
(4) Unsupervised learning clustering model-based
classification methods
Clustering is an unsupervised learning algorithm
that completely relies on the natural characteristics of
samples for identification. The basis of the clustering
concept is that for a given data set of M samples, a
given the number of clusters (K) (K<M) initializes the
category to which each sample belongs. Then,
iteration and reclassification of the data set according
to certain rules changes the class relationship between
samples and clusters, so that each new division is
better classified than the previous division (Zhang,
2019).
Current unsupervised learning clustering model-
based methods for clinical classification includes
Latent class analysis (LCA) cluster (Ning, 2019), K-
means (Arun, 2020), Meanshift (He, 2019),
hierarchical clustering (Laszlo, 2020), and scClustViz
(Brendan, 2018).
4 DISCUSSION
4.1 Problem Summaries
Although current classification methods can solve
some of the problems faced by clinical researchers,
because different authors of articles have a different
understanding of data sources, feature selection,
number of classification, classification methods, and
classification results evaluation, it is difficult to form
a consensus on the classification results. These results
lack interpretability and universality, which affects
the promotion and application of the classification
results.
(1) Data sources
In terms of data sources, because the data sources
used for classification are different, the data volume
and data quality of these data sources will also be
different (Wu, 2003). If the amount of data is small
and the quality of the data is poor, the credibility and
interpretability of the classification results will be
correspondingly reduced.
At the same time, most of the data used for clinical
classification is limited to the structured data from the
electronic medical record database, and there is a lack
of combined use of unstructured data and structured
data, such as text data (such as physical patient case
records) and image data (such as medical imaging
data), which may influence the credibility of the
classification results.
(2) Feature selection
Feature selection results have a direct and
significant impact on the classification results.
Because different researchers have different levels of
professional knowledge and understanding, current
feature selection methods cause different authors to
choose different features for classification on an
identical issue (Tian, 2021). At the same time, these
feature selection methods lack consideration of the
correlation among features. These issues cause
difficulties in reaching consensus with respect to
clinical classification results.
(3) Number of classification
It is also difficult to reach consensus to determine
the number of classification using current clinical
classification methods. Under normal circumstances,
most of the literature (CDC, 2020) will determine the
number of patient classifications based on the
patient's condition or certain disease characteristics,
and some authors will also perform secondary
classification using the initial classification results
(Zou, 2003). However, there is a lack of theoretical
support for the number of classification, resulting in
different authors using a different number of
classification for the same disease. For example, Wu
et al., (Wu, 2003) classified Severe Acute Respiratory
Syndrome (SARS) into ordinary, mild, severe, and
very severe, while Zou et al., (Zou, 2003) classified
SARS into ordinary, severe, and very severe.
Therefore, in this example, the question remains as to
whether SARS should be stratified into three or four
categories. Unfortunately, different articles fail to
explain the reasons for their chosen number of
classification in detail, and these articles can only
justify their own classifications, which also causes the
classification results to not be widely recognized.
(4) Selection of classification methods and
evaluation of classification results
Classification method was another important
factor that affected clinical classification results.
Different classification methods may generate
different classification results. Most clinical
Methods for Clinical Disease Classification and Clustering: A Review
119

classification results based on personal professional
knowledge are validated based on statistical analysis
(Monica, 2020; Emmanuel, 2018; Vincenzo, 2021).
Such classification evaluation can only explain the
rationality of the classification method, but does not
indicate whether there were other better classification
methods and results available.
At the same time, some classification models did
not compare the classification results with familiar
models, nor do they use credible classification
evaluation indicators to evaluate the classification
results. For example, Arun S et al., (Arun, 2020) only
used K-means for clustering; however, this paper did
not compare the results of the model with results of
other clustering models; meanwhile, the classification
results were not analyzed by using clustering effect
evaluation indicators, and the authors thus cannot
vouch for the credibility of their classification results.
4.2 Strategy
Taking the above problems into account, we propose
the following countermeasures to improve the
credibility and interpretability of classification
results.
(1) Data source
In order to improve the quantity and quality of
data, we recommended that researchers delete all
private data from patients and open source all the
original data. When discussing clinical classification,
researchers should also try to use open source data,
and use structured data and unstructured data at the
same time, so that the results of classification can be
recognized by more people. Current open source
medical data sets and databases include the breast-
cancer-Wisconsin data set, the Pima-Indians-data set,
the COVID-19 data set, and the Artificial Intelligence
Center of Stanford University for Medicine and
Imaging (AIMI) free repository of medical imaging
data sets, et al. At the same time, in order to improve
the quality of data, before classifying clinical data,
data preprocessing is required, including data
standardization, outlier and missing value processing,
and data integration (Ahady, 2021).
(2) Feature selection
We recommended using expert knowledge +
mathematical models to scientifically select clinical
classification features. The selection of clinical
classification features can use expert knowledge to
screen important clinical features first, and
subsequently the feature selection model to determine
the importance of the features to the classification
results, and finally the expert knowledge and feature
selection model to comprehensively decide which
features may be used for classification. This feature
selection method not only uses expert knowledge and
thus conforms to the public's expectation, but also
uses a mathematical model, which enhances the
scientific base of the classification feature selection
method, and the selected features are thus more easily
identified by professionals.
At the same time, there is another concept in the
feature dimensionality reduction model called feature
extraction, which uses existing features to combine
and generate new features (He, 2019; Li, 2019). For
example, CD4/CD8 ratio is a combined feature. This
combined feature has a certain relationship with non-
AIDS diseases (Cristina, 2015) and immune function
reconstruction (Jing, 2018), and has clinical
significance for its research. Therefore, the feature
extraction method can also be attempted in the feature
selection method.
(3) Number of classification and classification
model
The process of determining the number of
classification and the classification model should be
carried out at the same time. Firstly, based on expert
knowledge and the characteristics of the data used for
clinical classification, the scope of the number of
classification should be determined. Then the
appropriate classification model should be selected
based on the volume of data and the characteristics of
the data.
For supervised clinical data, typical supervised
learning models could be selected for clinical
classification (Jonathan, 2016), including logistic
regression, linear regression, support vector machine,
random forest, neural network, and so on.
For unsupervised clinical data (Zhang, 2019),
clustering algorithms could be used for classification.
If the amount of data is less than 1012 bytes, it can be
considered to be a small sample analysis. The
algorithms used included partition clustering (such as
K-means Clustering (Arun, 2020)), hierarchical
clustering (such as Agglomerative Hierarchical
Clustering (Laszlo, 2020)), artificial neural network
clustering (such as Self-Organizing Map), nuclear
clustering (such as Support Vector Clustering),
sequence data clustering (such as Trajectory
Clustering), etc. For massive unsupervised clinical
data, distributed clustering or parallel clustering may
be selected for clinical classification (Zhang, 2019).
If there is only a small amount of supervised
learning clinical data and a large amount of
unsupervised learning clinical data, a semi-
supervised learning model (Qin, 2019) could also be
used for clinical classification. Related methods
include Constraint-based Semi-supervised Clustering
ICBB 2022 - International Conference on Biotechnology and Biomedicine
120

(Wei, 2018), Distance-based Semi-supervised
Clustering (Yang, 2016), and Constraint and Distance
based Semi-supervised Clustering (Yu, 2014).
After classification is completed, the different
classification results need to be evaluated and
compared. Supervised learning models generally
used Precision, Recall, Accuracy, or F1-scores for
evaluation (He, 2019). If cross-validation was used to
prevent overfitting, then the cross-validation score is
used for evaluation. Unsupervised clustering models
generally use external indicators (such as the Jaccard
Coefficient, the Fowlkes and Mallows Index, and the
Rand Index) and internal indicators (such as the
Silhouette Coefficient, the Davies-Bouldin Index,
and the Dunn Index) to evaluate results (Johns, 2020).
The number of classification and the classification
models determined by the above methods are
supported by expert knowledge and mathematical
models, which can improve the underlying scientific
basis of the classification methods and the
interpretability of the classification results, and lead
to the classification results being recognized by more
medical professionals.
5 CONCLUSION
This review summarizes the general ideas of clinical
classification, the selection of clinical classification
features and classification methods, illustrates current
shortcomings of clinical classification, and proposes
corresponding countermeasures.
In the future, it is hoped that a large number of
data sets and more universal classification methods
are introduced to assist clinicians and scientific
researchers in the task of classification.
FUNDING
This study was supported by the Chongqing Science
and Technology Bureau Project (cstc2019jscx-
fxydX0037).
CONSENT FOR PUBLICATION
All authors have provided their individual consent for
publication of the manuscript.
COMPETING INTERESTS
None of the authors of this manuscript have
competing interests to declare.
ACKNOWLEDGEMENTS
We would like to express our gratitude to the funder
of this research.
AVAILABILITY OF DATA AND
MATERIAL
All data relevant to this study may be obtained from
Pubmed and CNKI.
REFERENCES
Ahady DH, Chen YJ, Leonard RD, Megahed FM,
JonesFarmer LA. Explaining Predictive Model
Performance: An Experimental Study of Data
Preparation and Model Choice. Big Data. 2021.10.
https://doi.org/10.1089/big.2021.0067.
Arcangelo P, Petr VT, Vincenzo P, Sergey E, et al.
Classifications and Clinical Assessment of
Haemorrhoids: The Proctologist's Corner. Reviews on
recent clinical trials. 2021; 16(1): 10-16.
https://doi.org/10.2174/1574887115666200312163940
.
Ahmad A, Ishan M, Sachin A, Patricia BM, et al. Machine
learning based classification and diagnosis of clinical
cardiomyopathies. Physiological genomics. 2020; 52:
1-42.
https://doi.org/10.1152/physiolgenomics.00063.2020.
Annabel LWG, Maayke MPK, Jelmer R, Asra G, et al.
Classification for treatment urgency for the
microphthalmia/anophthalmia spectrum using clinical
and biometrical characteristics. Acta ophthalmologica.
2020; 98(5): 514-520.
https://doi.org/10.1111/aos.14364.
Arun S, Peter AC, Gary C, Philip DC. Characterisation of
Upper Airway Collapse in OSA Patients Using Snore
Signals: A Cluster Analysis Approach. Annual
International Conference of the IEEE Engineering in
Medicine and Biology Society. 2020; 39(1): 5124-
5127.
https://doi.org/10.1109/EMBC44109.2020.9175591.
Asbjørn TB, Lars AN, Pascal M. Computer methods in
biomechanics and biomedical engineering. 2010; 13(6):
677-83. https://doi.org/10.1080/10255840903446979.
Artificial Intelligence Center of Stanford University.
https://stanfordaimi.azurewebsites.net.
Methods for Clinical Disease Classification and Clustering: A Review
121

Bernadette R, Prasad K. A Clinical Classification of
Pigmentary Disorders. Pigmentary Skin Disorders.
2018. 1-26.
Bi R, Jiang N, Yin Q, Chen H, et al. A new clinical
classification and treatment strategies for
temporomandibular joint ankylosis. International
journal of oral and maxillofacial surgery. 2020; 49(11):
1449-1458.
https://doi.org/10.1016/j.ijom.2020.02.020.
Brendan TI, Gary DB. scClustViz - Single-cell RNAseq
cluster assessment and visualization. F1000 Research.
2018; 7: ISCB Comm J-1522.
https://doi.org/10.12688/f1000research.16198.2.
Brian P, Michael R. Defining subgroups of patients with a
stiff and painful shoulder: an analytical model using
cluster analysis. Disability and rehabilitation. 2021;
43(4): 537-544.
https://doi.org/10.1080/09638288.2019.1631891.
Cristina M, Patrizia L, Alessandro CL, Giuseppe L, et al.
CD4/CD8 ratio normalisation and non-AIDS-related
events in individuals with HIV who achieve viral load
suppression with antiretroviral therapy: an
observational cohort study. The lancet. HIV. 2015;
2(3): e98-106. https://doi.org/10.1016/S2352-
3018(15)00006-5.
Chen YK, Wu H. Expert Consensus on Diagnosis and
Treatment of Pneumocystis Pneumonia in AIDS
Patients in China, Journal of Southwest
University(Natural Science Edition), 2020;42:49-60.
https://doi.org/10.13718/j.cnki.xdzk.2020.07.004.
China National Health Commission, China Center for
Disease Control and Prevention. COVID-19 Diagnosis
and Treatment Plan (Trial Version 8).
http://www.nhc.gov.cn/yzygj/s7653p/202008/0a7bdf1
2bd4b46e5bd28ca7f9a7f5e5a.shtml, 2020.8.18.
Dana P, Ji HJY, Gerard JB, John DO, et al. Identifying
subtypes of mild cognitive impairment in Parkinson's
disease using cluster analysis. Journal of neurology.
2020; 267(11): 3213-3222.
https://doi.org/10.1007/s00415-020-09977-z.
Daniele Cardaropoli, Myron Nevins, Paolo Casentini. A
Clinical Classification System for the Treatment of
Postextraction Sites. The International journal of
periodontics & restorative dentistry. 2021;41(2): 227-
232. https://doi.org/10.11607/prd.5069.
Emmanuel KA, Paul F, Mi YE, Soung MK, et al. Clinical
classification of cervical necrotizing fasciitis. European
archives of oto-rhino-laryngology: official journal of
the European Federation of Oto-Rhino-Laryngological
Societies (EUFOS): affiliated with the German Society
for Oto-Rhino-Laryngology - Head and Neck Surgery.
2018; 275(12): 3067-3073.
https://doi.org/10.1007/s00405-018-5155-5.
Elisa D, Antonio C, Geòrgia E, Jordi C, et al. Cluster
analysis of clinical data identifies fibromyalgia
subgroups. PLoS One. 2013; 8(9): e74873.
https://doi.org/10.1371/journal.pone.0074873.
Emma A, Petter S, Annemari K, Mats M. Novel subgroups
of adult-onset diabetes and their association with
outcomes: a data-driven cluster analysis of six
variables. The lancet. Diabetes & endocrinology. 2018;
6(5): 361-369. https://doi.org/10.1016/S2213-
8587(18)30051-2.
Fang XJ, Luo H, Lun YH, Zhu JQ. Genotyping of major
HIV-1 strains and its impact on drug resistance in
Dongguan. South China Journal of Preventive
Medicine. 2020; 46(3) :227-234. https://doi.org/10.
12183/j. scjpm. 2020. 0227.
He XQ. Python language programming and medical
application. China Railway Publishing House Co.,
LTD. 2019.12.
Huang LL, Zhang Y, Yang XM. AIDS patients with hepatic
tuberculosis clinical value of ultrasound to change the
image and classification. Imaging Research and
Medical Application, 2017; 15: 102-104.
Hua QC, Yang SJ, Niang BK, Feng XM, et al. The clinical
dialectical types and prospects of Tibetan medicine in
the treatment of chloasma. Chinese Journal of Health
and Nutrition. 2020; 30(28): 54.
José FBC, Marlene AJ, Joseph J. Functional gait disorders,
clinical phenomenology, and classification.
Neurological sciences: official journal of the Italian
Neurological Society and of the Italian Society of
Clinical Neurophysiology. 2020; 41(4): 911-915.
https://doi.org/10.1007/s10072-019-04185-8.
Jonathan B, Samah JF, Cynthia AB, Julie AW.
Classification of radiology reports for falls in an HIV
study cohort, Journal of the American Medical
Informatics Association. 2016; 23: 113-117.
https://doi.org/10.1093/jamia/ocv155.
Jessica JFW, Ingrid ES, Robert SF. The new definition and
classification of seizures and epilepsy. Epilepsy
research. 2018; 139: 73-79.
https://doi.org/10.1016/j.eplepsyres.2017.11.015.
Johns Hopkins University Center for Systems Science and
Engineering.
https://github.com/CSSEGISandData/COVID-19.
Jing FH, Lyu W, Li TS. A new view of CD4/Cd8 ratio as
an immune reconstitution marker in HIV-infected
individuals. Chinese Journal of AIDS & STD. 2018;
24(6): 643-644.
https://doi.org/10.13419/j.cnki.aids.2018.06.32.
Kubéraka M, Benjamin G, Damien A, Marguerite G, et al.
Development of a New Classification System for
Idiopathic Inflammatory Myopathies Based on Clinical
Manifestations and Myositis-Specific Autoantibodies.
JAMA neurology. 2018; 75(12): 1528-1537.
https://doi.org/10.1001/jamaneurol.2018.2598.
Keren MG, Tali BA, Samir N. Cluster analysis based
clinical profiling of Idiopathic Pulmonary Fibrosis
patients according to comorbidities evident prior to
diagnosis: a single-center observational study.
European journal of internal medicine. 2020; 80: 18-23.
https://doi.org/10.1016/j.ejim.2020.05.023.
Kaggle. https://www.kaggle.com/uciml/breast-cancer-
wisconsin-data.
Kaggle. https://www.kaggle.com/uciml/pima-indians-
diabetes-database.
Laszlo M, Karl WO, Gerd H, Richard S. Cluster Analysis
of Early Postnatal Biochemical Markers May Predict
ICBB 2022 - International Conference on Biotechnology and Biomedicine
122

Development of Retinopathy of Prematurity.
Translational vision science & technology. 2020; 9(13):
14. https://doi.org/10.1167/tvst.9.13.14.
Leila A, Zahra RS, Mohsen R, Hadi RS. Kinematic cluster
analysis of the crouch gait pattern in children with
spastic diplegic cerebral palsy using sparse K-means
method. Clinical biomechanics (Bristol, Avon). 2021;
81: 105248.
https://doi.org/10.1016/j.clinbiomech.2020.105248.
Liu M, Liu JF, Wu B. Progress and interpretation of
classification and classification of cerebrovascular
diseases. Chinese Journal of Neurology. 2017; 50(3):
163-167.
Li ZQ, Du JQ, Nie B, et al. Summary of feature selection
methods. Computer Engineering and Applications.
2019, 55(24):10-19.
https://doi.org/10.3778/j.issn.1002-8331.1909-0066.
Li J, Xiang F. Identification of risk factors for coronary
heart disease and establishment of their prediction
model. Chinese Journal of Medical Library and
Information. 2020; 29(6): 7-13.
https://doi.org/10.3969/j.issn.1671-3982.2020.06.002.
Ma XD, Michael NG, Xu S, Xu ZM, et al. Development
and validation of prognosis model of mortality risk in
patients with COVID-19. Epidemiology and infection.
2020; 148: e168.
https://doi.org/10.1017/S0950268820001727.
Miller BS, Turcu AF, Nanba AT, Hughes DT, et al.
Refining the Definitions of Biochemical and Clinical
Cure for Primary Aldosteronism Using the Primary
Aldosteronism Surgical Outcome (PASO)
Classification System. World journal of surgery. 2018;
42(2): 453-463. https://doi.org/10.1007/s00268-017-
4311-1.
Monica HV, Antonio C, Ambar K, David MO. A clinical
classification system for grading platinum
hypersensitivity reactions. Gynecologic oncology.
2020; 159(3): 794-798.
https://doi.org/10.1016/j.ygyno.2020.09.009.
Ning CX, Chen XX, Lin HJ, Qiao XT, et al. Characteristics
of sleep disorder in HIV positive and HIV negative
individuals: a cluster analysis. Chinese Journal of
Epidemiology. 2019; 40(5): 499-504.
https://doi.org/10.3760/cma.j.issn.0254-
6450.2019.05.002.
Noor D, Louise CS, Andrew VK, Celia C, et al.
Identification of a 251 gene expression signature that
can accurately detect M. tuberculosis in patients with
and without HIV co-infection. PLoS One. 2014; 9(2):
e89925. https://doi.org/10.1371/journal.pone.0089925.
Paweł C, Aldona P, Jacek G, Grzegorz K, Dorota K.
Psoriatic arthritis – classification, diagnostic and
clinical aspects. Dermatol Rev/Przegl Dermatol 2020;
107: 32-43.
https://doi.org/https://doi.org/10.5114/dr.2020.93969.
Pranab H, Ian DP, Dominic ES, Michael AB, et al. Cluster
analysis and clinical asthma phenotypes. American
journal of respiratory and critical care medicine. 2008;
178(3): 218-224. https://doi.org/10.1164/rccm.200711-
1754OC.
Qin Y, Ding SF. Survey of Semi-supervised Clustering.
Computer Science. 2019; 46(9): 15-21. https://doi.org/1
0.11 8 9 6/j.issn.1002-137X.2019.09.002.
Sandra A, Maria C, Barbara H, Anne W, et al. A
mechanistic classification of clinical phenotypes in
neuroblastoma. Science. 2018; 362(6419): 1165-1170.
https://doi.org/10.1126/science.aat6768.
Shuai Y, Song TL, Wang JP, Shen H. Research of
Equipment Support Data Preparation Methods. Fire
Control & Command Control. 2018, 43(09):135-139.
https://doi.org/10.3969/j.issn.1002-0640.2018.09.028.
Sun LP, Zhang LJ. Clinical big data analysis and mining.
Publishing House of Electronics Industry. 2020.11.
Sun JJ, Liu Y. Interpretation of Guidelines for Clinical
Classification and Surgical Classification of Otitis
Media (2012). Chinese Journal of Otorhinolaryngology
Head and Neck Surgery. 2013; 48(1): 6-10.
https://doi.org/10.3760/cma.j.issn.1673-
0860.2013.01.004.
Szmulewicz A, Millett CE, Shanahan M, Gunning FM,
Burdick KE. Emotional processing subtypes in bipolar
disorder: A cluster analysis. Journal of affective
disorders. 20201; 266: 194-200.
https://doi.org/10.1016/j.jad.2020.01.082.
Shi HF, Liu Q. Application of management of case
classification in the analysis of medical cost of medical
insurance case. Modern Preventive Medicine. 2010;
37(6): 1055-105.
Thijs WCT, Antien LM, Kerstin A, Arthur HC, et al.
Pathologic classification of diabetic nephropathy.
Journal of the American Society of Nephrology: JASN.
2010; 21(4): 556-63.
https://doi.org/10.1681/ASN.2010010010.
Tian LL. Clinical Study on TCM Syndrome Differentiation
after Chemotherapy for Advanced Triple Negative
Breast Cancer. Chinese Remedies & Clinics. 2021;
21(3): 444-446.
https://doi.org/10.11655/zgywylc2021.03.037.
Tsang JYS, Tse GM. Molecular Classification of Breast
Cancer. Adv Anat Pathol. 2020; 27(1):27-35.
https://doi.org/10.1097/PAP.0000000000000232.
Tilahun NH, Frederick MW, Shukri FM, Martin KM, et al.
Patterns of non-communicable disease and injury risk
factors in Kenyan adult population: a cluster analysis.
BMC Public Health. 2018; 18(Suppl 3): 1225.
https://doi.org/10.1186/s12889-018-6056-7.
Varol BA, Supriya N, Mobashir HS, Joanna F, et al.
Classification of Decompensated Heart Failure from
Clinical and Home Ballistocardiography. IEEE
transactions on bio-medical engineering. 2020; 67(5):
1303-1313.
https://doi.org/10.1109/TBME.2019.2935619.
Viprakasit V, Ekwattanakit S. Clinical Classification,
Screening and Diagnosis for Thalassemia.
Hematology/oncology clinics of North America. 2018;
32(2): 193-211.
https://doi.org/10.1016/j.hoc.2017.11.006.
Vincenzo L, Francesca N, Roser P, Serena G. Parkinsonism
in children: Clinical classification and etiological
spectrum. Parkinsonism & related disorders. 2021; 82:
Methods for Clinical Disease Classification and Clustering: A Review
123

150-157.
https://doi.org/10.1016/j.parkreldis.2020.10.002.
Wang QC, Wang Y, Wang JC, Wang YB. Clinical
classification of clival chordomas for transnasal
approaches. Neurosurgical review. 2020; 43(4): 1201-
1210. https://doi.org/10.1007/s10143-019-01153-w.
Wang CY, Guo L, Wang XC, Wu Y. Analysis of risk
factors for death of patients with severe pneumonia.
Medical Journal of Wuhan University. 2020; 41(1):
110-113. https://doi.org/10.14188/j.1671‐
8852.2018.1213.
Wei ST, Li ZX, Zhang CL. Combined constraint-based
with metric-based in semi-supervised clustering
ensemble. International Journal of Machine Learning
and Cybernetics. 2018; 9(7): 1085-1100.
https://doi.org/10.1007/s13042-016-0628-6.
William P, Cheshire J. Clinical classification of orthostatic
hypotensions. Clinical autonomic research: official
journal of the Clinical Autonomic Research Society.
2017; 27(3): 133-135. https://doi.org/10.1007/s10286-
017-0414-x.
Wu H, Chen XY, Zhao CH. Initial study of clinical
classification and staging in severe acute respiratory
syndrome, Chin J infect Dis, 2003;21:176-179.
Xu XM, Zhang YY, Du PC, Li M, et al. MLST
classification and Clinical Characteristics of New
Crypticoccus Infections in AIDS Partitions. Labeled
Immunoassays and Clinical, 2020;27:829-832.
Xie ZP, Zhu B. Correlation between CT Manifestations:
Types and Prognosis of Pneumocystis Pneumonia in
AIDS Patients, Journal of Clinical Radiology,
2018;37:228-232.
Xiang Q, Chen YP. Standardization method for
hospitalization cost analysis based on distribution of
disease-case classification and operation. Chinese
Hospital Management. 2009;29(5): 31-34.
https://doi.org/10.3969/j.issn.1001-5329.2009.05.015
Yang J, Deng T. A semi-supervised multiview spectral
clustering algorithm based on distance metric learning.
Journal of Sichuan University (Engineering Science
Edition). 2016; 48(1): 146-151.
https://doi.org/10.15961/j.jsuese.2016.01.022.
Yuan J, Deng CG, Li QS, Yu Q, et al. Retrospective
analysis of prognostic factors in 289 AIDS patients
complicated with severe Pneumocystis pneumonia.
Retrospective analysis of prognostic factors in 289
AIDS patients complicated with severe Pneumocystis
pneumonia. Chinese Journal of Infection and
Chemotherapy. 2020; 20(6): 594-600.
https://doi.org/10.16718/j.1009-7708.2020.06.002.
Yu ZW, Chen HS, You JN, Wong HS, Liu JM, Li L, et al.
Double Selection Based Semi-Supervised Clustering
Ensemble for Tumor Clustering from Gene Expression
Profiles. IEEE/ACM Trans Comput Biol Bioinform.
2014; 11(4):7 27-40.
https://doi.org/10.1109/TCBB.2014.2315996.
Yulia L, Olga B, Alexander N, Svetlana A, et al. Clinical
Classification of Arrhythmogenic Right Ventricular
Cardiomyopathy. Pulse (Basel, Switzerland). 2020;
8(1-2): 21-30. https://doi.org/10.1159/000505652.
Yun JL, Haeng JL, Seong JK. Clinical Features of Duane
Retraction Syndrome: A New Classification. Korean
journal of ophthalmology: KJO. 2020; 34(2): 158-165.
https://doi.org/10.3341/kjo.2019.0100.
Yang H, Li SG, Xiang X, Lv Y, et al. Clinical classification
and individualized design for the treatment of
basicranial artery injuries. Medicine (Baltimore). 2019;
98(11): e14732.
https://doi.org/10.1097/MD.0000000000014732.
Zou ZS, Yang YP, Chen JM, Xin SJ, et al. Features of
clinical stages and types of severe acute respiratory
syndrome and their clinical significance. Journal of
PLA Medical, 2003; 28: 777-780.
Zhang YL, Zhou YJ. Review of clustering algorithms.
Journal of Computer Applications. 2019;39(7): 1869-
1882.
ICBB 2022 - International Conference on Biotechnology and Biomedicine
124
